Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
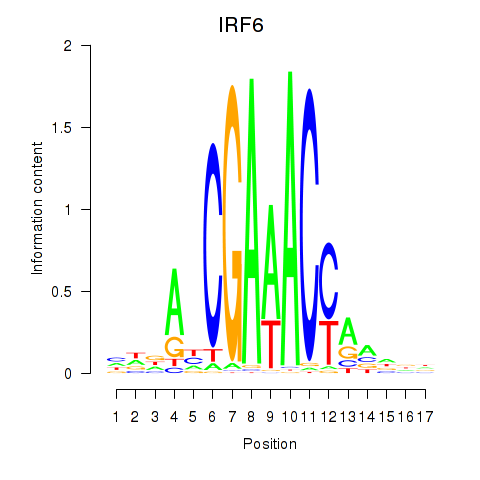
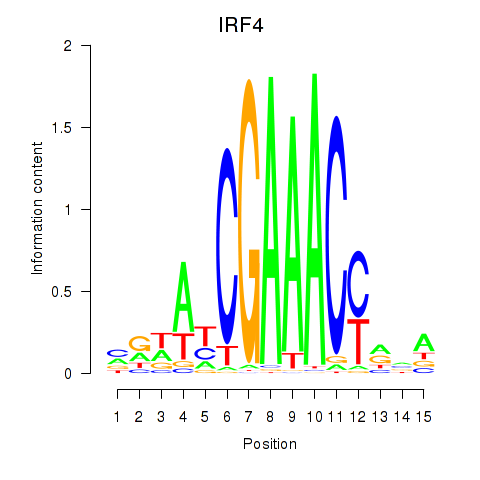
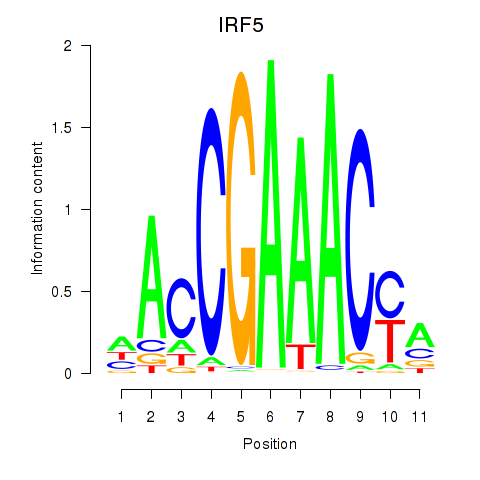
Results for IRF6_IRF4_IRF5
Z-value: 6.08
Transcription factors associated with IRF6_IRF4_IRF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF6
|
ENSG00000117595.6 | interferon regulatory factor 6 |
|
IRF4
|
ENSG00000137265.10 | interferon regulatory factor 4 |
|
IRF5
|
ENSG00000128604.14 | interferon regulatory factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF5 | hg19_v2_chr7_+_128577972_128578047 | -0.68 | 1.4e-01 | Click! |
| IRF6 | hg19_v2_chr1_-_209979465_209979494 | -0.21 | 6.9e-01 | Click! |
| IRF4 | hg19_v2_chr6_+_391739_391759 | 0.13 | 8.0e-01 | Click! |
Activity profile of IRF6_IRF4_IRF5 motif
Sorted Z-values of IRF6_IRF4_IRF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_615942 | 44.69 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr21_+_42798094 | 28.19 |
ENST00000398598.3
ENST00000455164.2 ENST00000424365.1 |
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr21_+_42792442 | 27.91 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr9_-_32526184 | 24.40 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr9_-_32526299 | 23.54 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr1_+_948803 | 19.99 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr21_+_42798124 | 17.97 |
ENST00000417963.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr21_+_42798158 | 17.92 |
ENST00000441677.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr11_-_615570 | 16.74 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr12_+_113344755 | 15.63 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr21_+_42797958 | 15.42 |
ENST00000419044.1
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr12_+_113344582 | 15.30 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_-_321050 | 14.35 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr3_-_146262365 | 14.20 |
ENST00000448787.2
|
PLSCR1
|
phospholipid scramblase 1 |
| chr12_+_113344811 | 13.65 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_122283079 | 13.44 |
ENST00000471785.1
ENST00000466126.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_122283100 | 13.01 |
ENST00000492382.1
ENST00000462315.1 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_122283424 | 12.11 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_-_146262352 | 11.13 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr2_-_37384175 | 10.49 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr3_-_146262428 | 10.49 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_146262293 | 10.49 |
ENST00000448205.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_146262488 | 9.08 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr12_+_113376157 | 8.40 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr3_-_146262637 | 8.28 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr9_-_100881466 | 8.12 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr12_+_113376249 | 8.02 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr14_+_94577074 | 7.69 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr3_+_122283064 | 7.45 |
ENST00000296161.4
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr3_+_122283175 | 7.32 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr2_-_7005785 | 6.70 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr20_-_62203808 | 5.41 |
ENST00000467148.1
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr2_-_191878681 | 5.21 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr2_-_191878874 | 5.00 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr19_+_10196981 | 4.91 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_57335280 | 4.62 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr2_-_163175133 | 4.60 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr11_-_321340 | 4.05 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chrM_+_9207 | 3.66 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr14_-_102552659 | 3.53 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr19_-_17516449 | 3.23 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr4_+_89378261 | 3.06 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr10_+_91174314 | 2.78 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr15_-_45670717 | 2.68 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr5_+_178286925 | 2.67 |
ENST00000322434.3
|
ZNF354B
|
zinc finger protein 354B |
| chr2_-_55920952 | 2.50 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr12_-_62653903 | 2.44 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chrX_+_23801280 | 2.41 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr19_+_10196781 | 2.15 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr8_+_40018977 | 2.04 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr15_-_75748143 | 2.00 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr7_-_33080506 | 1.97 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr3_-_182880541 | 1.96 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr7_+_12250833 | 1.96 |
ENST00000396668.3
|
TMEM106B
|
transmembrane protein 106B |
| chr4_+_144258288 | 1.87 |
ENST00000514639.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr10_+_91061712 | 1.76 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr9_+_111696664 | 1.75 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr3_-_180707306 | 1.73 |
ENST00000479269.1
|
DNAJC19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr3_-_96337000 | 1.68 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr18_+_22006646 | 1.64 |
ENST00000585067.1
ENST00000578221.1 |
IMPACT
|
impact RWD domain protein |
| chr4_+_95373037 | 1.57 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr1_-_238108575 | 1.57 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr7_-_77045617 | 1.53 |
ENST00000257626.7
|
GSAP
|
gamma-secretase activating protein |
| chr9_-_103115135 | 1.52 |
ENST00000537512.1
|
TEX10
|
testis expressed 10 |
| chr9_+_95820966 | 1.51 |
ENST00000375472.3
ENST00000465709.1 |
SUSD3
|
sushi domain containing 3 |
| chr20_-_55934878 | 1.50 |
ENST00000543500.1
|
MTRNR2L3
|
MT-RNR2-like 3 |
| chr6_+_89791507 | 1.49 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr17_-_54991395 | 1.48 |
ENST00000316881.4
|
TRIM25
|
tripartite motif containing 25 |
| chr11_-_57334732 | 1.47 |
ENST00000526659.1
ENST00000527022.1 |
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr10_+_91174486 | 1.44 |
ENST00000416601.1
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr17_-_54991369 | 1.40 |
ENST00000537230.1
|
TRIM25
|
tripartite motif containing 25 |
| chr1_-_54411255 | 1.39 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr5_+_68788594 | 1.34 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr5_-_79946820 | 1.32 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr6_-_86353510 | 1.32 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr11_-_10530723 | 1.32 |
ENST00000536684.1
|
MTRNR2L8
|
MT-RNR2-like 8 |
| chr12_+_69004805 | 1.31 |
ENST00000541216.1
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr7_-_95064264 | 1.30 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr6_-_26285737 | 1.28 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr6_+_53794780 | 1.28 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr12_-_56753858 | 1.24 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr19_+_12273866 | 1.22 |
ENST00000425827.1
ENST00000439995.1 ENST00000343979.4 ENST00000398616.2 ENST00000418338.1 |
ZNF136
|
zinc finger protein 136 |
| chr12_+_8850277 | 1.22 |
ENST00000539923.1
ENST00000537189.1 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr17_+_6659153 | 1.21 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr1_-_52344416 | 1.19 |
ENST00000544028.1
|
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr7_-_138794394 | 1.16 |
ENST00000242351.5
ENST00000471652.1 |
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr3_-_113464906 | 1.16 |
ENST00000477813.1
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr12_+_69004619 | 1.16 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr5_+_138678131 | 1.14 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_+_179262905 | 1.14 |
ENST00000539888.1
ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1
|
sterol O-acyltransferase 1 |
| chr5_+_118668846 | 1.13 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr18_-_812517 | 1.13 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr19_+_36705504 | 1.12 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr22_+_18632666 | 1.10 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr12_+_79258547 | 1.06 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr6_-_100016678 | 1.06 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr3_+_159706537 | 1.04 |
ENST00000305579.2
ENST00000480787.1 ENST00000466512.1 |
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr18_+_22006580 | 1.04 |
ENST00000284202.4
|
IMPACT
|
impact RWD domain protein |
| chr5_-_95297534 | 1.03 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr12_+_79258444 | 1.02 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr6_+_64282447 | 1.01 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr18_-_51884204 | 1.00 |
ENST00000577499.1
ENST00000584040.1 ENST00000581310.1 |
STARD6
|
StAR-related lipid transfer (START) domain containing 6 |
| chr5_-_159846066 | 0.99 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr8_+_107630340 | 0.99 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr11_+_5710919 | 0.98 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr15_+_74287035 | 0.98 |
ENST00000395132.2
ENST00000268059.6 ENST00000354026.6 ENST00000268058.3 ENST00000565898.1 ENST00000569477.1 ENST00000569965.1 ENST00000567543.1 ENST00000436891.3 ENST00000435786.2 ENST00000564428.1 ENST00000359928.4 |
PML
|
promyelocytic leukemia |
| chr12_+_49760639 | 0.96 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr15_-_52263937 | 0.92 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chrM_+_10053 | 0.92 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_-_104119528 | 0.90 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr7_-_138794081 | 0.90 |
ENST00000464606.1
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr2_+_120770645 | 0.88 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr20_-_20033052 | 0.87 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr4_-_129208030 | 0.87 |
ENST00000503872.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr9_-_103115185 | 0.86 |
ENST00000374902.4
|
TEX10
|
testis expressed 10 |
| chr14_-_45603657 | 0.86 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr4_-_83295296 | 0.85 |
ENST00000507010.1
ENST00000503822.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr7_+_12250886 | 0.85 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr12_+_62654119 | 0.85 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr18_-_812231 | 0.84 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr2_+_85811525 | 0.83 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chrX_-_77041685 | 0.82 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr3_+_172468505 | 0.81 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr17_+_22022437 | 0.81 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr19_-_9649303 | 0.81 |
ENST00000253115.2
|
ZNF426
|
zinc finger protein 426 |
| chr13_+_51483814 | 0.80 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr1_-_169680745 | 0.80 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr5_-_133706695 | 0.79 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr7_-_98467629 | 0.79 |
ENST00000339375.4
|
TMEM130
|
transmembrane protein 130 |
| chr17_-_26903900 | 0.78 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr3_+_88199099 | 0.77 |
ENST00000486971.1
|
C3orf38
|
chromosome 3 open reading frame 38 |
| chr2_-_198299726 | 0.76 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr4_+_144257915 | 0.75 |
ENST00000262995.4
|
GAB1
|
GRB2-associated binding protein 1 |
| chr4_-_118006697 | 0.74 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr19_+_11466167 | 0.73 |
ENST00000591608.1
|
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr7_+_151038850 | 0.73 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr3_-_108308241 | 0.72 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr2_+_113299990 | 0.70 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr1_-_154580616 | 0.70 |
ENST00000368474.4
|
ADAR
|
adenosine deaminase, RNA-specific |
| chr4_-_83295103 | 0.69 |
ENST00000313899.7
ENST00000352301.4 ENST00000509107.1 ENST00000353341.4 ENST00000541060.1 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr22_+_36044411 | 0.68 |
ENST00000409652.4
|
APOL6
|
apolipoprotein L, 6 |
| chr2_+_32390925 | 0.68 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr12_-_51663959 | 0.68 |
ENST00000604188.1
ENST00000398453.3 |
SMAGP
|
small cell adhesion glycoprotein |
| chr14_+_105266933 | 0.68 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr7_+_106809406 | 0.66 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr8_+_19536083 | 0.66 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr2_-_99797473 | 0.65 |
ENST00000409107.1
ENST00000289359.2 |
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr1_-_52344471 | 0.65 |
ENST00000352171.7
ENST00000354831.7 |
NRD1
|
nardilysin (N-arginine dibasic convertase) |
| chr5_-_76788024 | 0.64 |
ENST00000515253.1
ENST00000414719.2 ENST00000507654.1 ENST00000514559.1 ENST00000511791.1 |
WDR41
|
WD repeat domain 41 |
| chr2_-_197664366 | 0.64 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr2_-_111435610 | 0.63 |
ENST00000447014.1
ENST00000420328.1 ENST00000535254.1 ENST00000409311.1 ENST00000302759.6 |
BUB1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr12_-_122711968 | 0.63 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr14_+_39736582 | 0.62 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr4_-_11431188 | 0.61 |
ENST00000510712.1
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chrX_-_119077695 | 0.61 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr2_+_120770686 | 0.60 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr10_+_105881906 | 0.60 |
ENST00000369727.3
ENST00000336358.5 |
SFR1
|
SWI5-dependent recombination repair 1 |
| chr20_+_48789121 | 0.59 |
ENST00000411453.1
|
RP11-112L6.4
|
RP11-112L6.4 |
| chr12_-_31477072 | 0.59 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr19_-_9649253 | 0.59 |
ENST00000593003.1
|
ZNF426
|
zinc finger protein 426 |
| chr4_+_86396265 | 0.59 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr11_-_85522172 | 0.58 |
ENST00000316356.4
ENST00000389960.4 ENST00000527523.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr11_-_47736896 | 0.58 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr9_+_102668915 | 0.58 |
ENST00000259400.6
ENST00000531035.1 ENST00000525640.1 ENST00000534052.1 ENST00000526607.1 |
STX17
|
syntaxin 17 |
| chr11_-_2162162 | 0.58 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr8_+_82066514 | 0.57 |
ENST00000519412.1
ENST00000521953.1 |
RP11-1149M10.2
|
RP11-1149M10.2 |
| chr18_+_52385068 | 0.57 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr3_+_52448539 | 0.57 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr16_-_3068171 | 0.55 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr2_-_99797390 | 0.54 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr4_+_86396321 | 0.54 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_+_217277271 | 0.54 |
ENST00000425815.1
|
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr3_-_142166796 | 0.54 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr5_-_61699698 | 0.53 |
ENST00000506390.1
ENST00000199320.4 |
DIMT1
|
DIM1 dimethyladenosine transferase 1 homolog (S. cerevisiae) |
| chr14_+_90528109 | 0.53 |
ENST00000282146.4
|
KCNK13
|
potassium channel, subfamily K, member 13 |
| chr14_+_89029336 | 0.53 |
ENST00000556945.1
ENST00000556158.1 ENST00000557607.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr4_-_104119488 | 0.51 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr12_+_54402790 | 0.51 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr1_-_25558963 | 0.50 |
ENST00000354361.3
|
SYF2
|
SYF2 pre-mRNA-splicing factor |
| chr6_-_160209471 | 0.48 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr12_+_75874460 | 0.48 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr11_-_2162468 | 0.48 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr17_+_40118773 | 0.47 |
ENST00000472031.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr7_-_19748640 | 0.47 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr7_-_98467543 | 0.46 |
ENST00000345589.4
|
TMEM130
|
transmembrane protein 130 |
| chr12_-_51664058 | 0.46 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr6_-_82462425 | 0.45 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr19_-_3029011 | 0.45 |
ENST00000590536.1
ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr7_-_107883678 | 0.45 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr8_-_101962777 | 0.45 |
ENST00000395951.3
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr14_+_89029253 | 0.44 |
ENST00000251038.5
ENST00000359301.3 ENST00000302216.8 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr3_+_118892411 | 0.44 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr7_-_130598059 | 0.44 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr9_+_112542572 | 0.43 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr2_+_217277137 | 0.43 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr4_-_103748271 | 0.43 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_+_56205878 | 0.42 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr14_-_103989033 | 0.41 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chrX_-_85302531 | 0.41 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr7_-_98467489 | 0.40 |
ENST00000416379.2
|
TMEM130
|
transmembrane protein 130 |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF6_IRF4_IRF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.5 | 61.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 13.1 | 52.5 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 6.4 | 63.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 5.0 | 104.8 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 3.7 | 40.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 2.6 | 10.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 2.4 | 29.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.7 | 6.7 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 1.4 | 16.4 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 1.2 | 36.5 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.9 | 2.7 | GO:0071264 | regulation of eIF2 alpha phosphorylation by amino acid starvation(GO:0060733) regulation of translational initiation in response to starvation(GO:0071262) positive regulation of translational initiation in response to starvation(GO:0071264) |
| 0.8 | 2.5 | GO:2000625 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.8 | 2.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.7 | 3.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.5 | 41.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.5 | 1.5 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.5 | 1.5 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.5 | 1.9 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 2.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.3 | 2.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.3 | 15.7 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.3 | 0.8 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.3 | 8.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.2 | 2.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 1.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 3.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 0.5 | GO:0051808 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 1.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.6 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 2.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 1.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 1.0 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 2.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 5.9 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 2.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 1.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.3 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 2.0 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.6 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.2 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 1.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 2.3 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 0.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 1.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 1.0 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.1 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 2.8 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.7 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.6 | GO:0040013 | negative regulation of locomotion(GO:0040013) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 1.6 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0050748 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 2.1 | GO:0048806 | genitalia development(GO:0048806) |
| 0.0 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 2.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 1.1 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.8 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.8 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0032268 | regulation of cellular protein metabolic process(GO:0032268) |
| 0.0 | 0.7 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.3 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 56.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 2.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 1.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 0.8 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 112.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 3.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 2.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 1.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 64.6 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.0 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 69.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 9.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 70.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 8.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 47.9 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 61.0 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 3.0 | 63.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.7 | 10.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.7 | 6.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.3 | 39.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 1.2 | 15.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.9 | 9.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.8 | 2.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.6 | 1.9 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.5 | 67.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.4 | 1.3 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.4 | 61.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.4 | 1.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 2.0 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.3 | 1.0 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.3 | 1.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 1.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.3 | 0.3 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.3 | 3.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.3 | 0.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 7.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 104.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 2.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 2.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 3.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 3.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) annealing activity(GO:0097617) |
| 0.1 | 6.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 13.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.3 | GO:0051880 | 5'-3' exoribonuclease activity(GO:0004534) G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 2.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0008124 | phenylalanine 4-monooxygenase activity(GO:0004505) 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.0 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 63.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 12.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 8.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 116.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 2.5 | 223.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 9.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 6.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 2.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 10.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 11.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 5.9 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |