Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
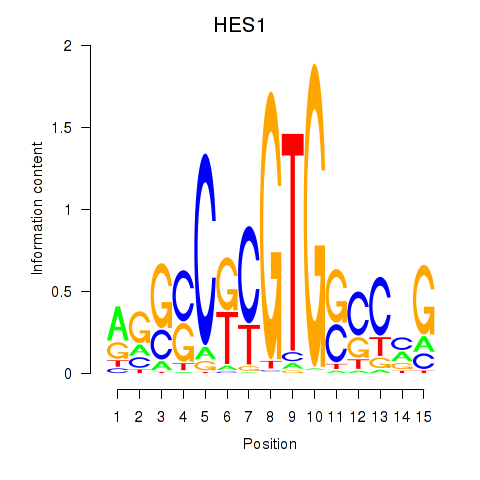
Results for HES1
Z-value: 0.88
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.3 | hes family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg19_v2_chr3_+_193853927_193853944 | -0.58 | 2.2e-01 | Click! |
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_94577074 | 1.10 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr11_-_615570 | 0.81 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr14_+_103394963 | 0.48 |
ENST00000559525.1
ENST00000559789.1 |
AMN
|
amnion associated transmembrane protein |
| chr2_+_86116396 | 0.43 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr8_+_38614754 | 0.39 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr14_+_67708344 | 0.33 |
ENST00000557237.1
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr10_+_104474353 | 0.33 |
ENST00000602647.1
ENST00000602439.1 ENST00000602764.1 |
SFXN2
|
sideroflexin 2 |
| chr15_+_89182156 | 0.30 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr10_-_95360983 | 0.29 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr3_+_121554046 | 0.29 |
ENST00000273668.2
ENST00000451944.2 |
EAF2
|
ELL associated factor 2 |
| chr15_+_89182178 | 0.28 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr2_+_187350973 | 0.28 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr8_+_22298767 | 0.27 |
ENST00000522000.1
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chrX_+_21958814 | 0.27 |
ENST00000379404.1
ENST00000415881.2 |
SMS
|
spermine synthase |
| chr2_-_100106419 | 0.26 |
ENST00000393445.3
ENST00000258428.3 |
REV1
|
REV1, polymerase (DNA directed) |
| chr8_-_99955042 | 0.25 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr22_+_24819565 | 0.25 |
ENST00000424232.1
ENST00000486108.1 |
ADORA2A
|
adenosine A2a receptor |
| chr19_-_7936344 | 0.23 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr2_+_232575168 | 0.23 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr20_-_3154162 | 0.23 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr1_+_53793885 | 0.22 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr7_-_155089251 | 0.22 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr19_-_3869012 | 0.21 |
ENST00000592398.1
ENST00000262961.4 ENST00000439086.2 |
ZFR2
|
zinc finger RNA binding protein 2 |
| chr19_+_843314 | 0.21 |
ENST00000544537.2
|
PRTN3
|
proteinase 3 |
| chr16_+_318638 | 0.20 |
ENST00000412541.1
ENST00000435035.1 |
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr17_-_48207115 | 0.20 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr22_+_24115000 | 0.20 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr2_+_46926326 | 0.19 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr8_-_142318398 | 0.19 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr22_+_29168652 | 0.19 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr12_-_25783684 | 0.19 |
ENST00000538178.1
|
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr16_+_2039946 | 0.19 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr9_-_107754034 | 0.19 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr17_-_80291818 | 0.19 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr9_+_133320301 | 0.19 |
ENST00000352480.5
|
ASS1
|
argininosuccinate synthase 1 |
| chr15_+_89181974 | 0.18 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr21_-_40685536 | 0.18 |
ENST00000341322.4
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr12_+_7022909 | 0.18 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr1_-_118472171 | 0.18 |
ENST00000369442.3
|
GDAP2
|
ganglioside induced differentiation associated protein 2 |
| chr4_+_57774042 | 0.18 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr4_-_10458982 | 0.18 |
ENST00000326756.3
|
ZNF518B
|
zinc finger protein 518B |
| chr22_-_42310570 | 0.18 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr22_+_29279552 | 0.17 |
ENST00000544604.2
|
ZNRF3
|
zinc and ring finger 3 |
| chr2_-_224702257 | 0.17 |
ENST00000409375.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr12_-_117537240 | 0.17 |
ENST00000392545.4
ENST00000541210.1 ENST00000335209.7 |
TESC
|
tescalcin |
| chr6_+_86159821 | 0.17 |
ENST00000369651.3
|
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr17_+_5973740 | 0.17 |
ENST00000576083.1
|
WSCD1
|
WSC domain containing 1 |
| chr1_+_6845384 | 0.17 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr11_+_64004888 | 0.17 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_-_9189144 | 0.17 |
ENST00000414642.2
|
GPR157
|
G protein-coupled receptor 157 |
| chr20_-_20693131 | 0.17 |
ENST00000202677.7
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr4_+_76439649 | 0.17 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr11_-_72852320 | 0.16 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr3_+_23851928 | 0.16 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr2_+_208576259 | 0.16 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr19_+_8274185 | 0.16 |
ENST00000558268.1
ENST00000558331.1 |
CERS4
|
ceramide synthase 4 |
| chr21_-_18985230 | 0.16 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr17_-_19880992 | 0.16 |
ENST00000395536.3
ENST00000576896.1 ENST00000225737.6 |
AKAP10
|
A kinase (PRKA) anchor protein 10 |
| chr8_+_22298578 | 0.16 |
ENST00000240139.5
ENST00000289963.8 ENST00000397775.3 |
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr11_-_111383064 | 0.16 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr12_+_49761147 | 0.15 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr4_-_185655212 | 0.15 |
ENST00000541971.1
|
MLF1IP
|
centromere protein U |
| chr2_-_200715834 | 0.15 |
ENST00000420128.1
ENST00000416668.1 |
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr2_-_96811170 | 0.15 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr14_+_93260642 | 0.15 |
ENST00000355976.2
|
GOLGA5
|
golgin A5 |
| chr1_+_220267429 | 0.14 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr1_-_101491319 | 0.14 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr14_+_102027688 | 0.14 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr2_+_187350883 | 0.14 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr12_+_107349606 | 0.14 |
ENST00000547242.1
ENST00000551489.1 ENST00000550344.1 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr1_-_45805607 | 0.14 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr14_+_56046990 | 0.14 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr18_+_74207477 | 0.14 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chrX_-_153095813 | 0.13 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr17_+_28256874 | 0.13 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr19_-_1652575 | 0.13 |
ENST00000587235.1
ENST00000262965.5 |
TCF3
|
transcription factor 3 |
| chrX_-_106243294 | 0.13 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr13_-_31039375 | 0.13 |
ENST00000399494.1
|
HMGB1
|
high mobility group box 1 |
| chr7_-_99774945 | 0.13 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr2_-_224702270 | 0.13 |
ENST00000396654.2
ENST00000396653.2 ENST00000423110.1 ENST00000443700.1 |
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr4_-_129209221 | 0.13 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr6_-_74233480 | 0.13 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr4_-_71705027 | 0.13 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr11_-_64511789 | 0.13 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_-_47546220 | 0.13 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr13_+_42031679 | 0.13 |
ENST00000379359.3
|
RGCC
|
regulator of cell cycle |
| chr19_-_59066327 | 0.13 |
ENST00000596708.1
ENST00000601220.1 ENST00000597848.1 |
CHMP2A
|
charged multivesicular body protein 2A |
| chr17_+_78075324 | 0.13 |
ENST00000570803.1
|
GAA
|
glucosidase, alpha; acid |
| chrX_-_54070388 | 0.13 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr20_-_61733657 | 0.13 |
ENST00000608031.1
ENST00000447910.2 |
HAR1B
|
highly accelerated region 1B (non-protein coding) |
| chr6_-_114292284 | 0.13 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr6_-_114292449 | 0.12 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr5_+_63461642 | 0.12 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr19_+_8274512 | 0.12 |
ENST00000560412.1
|
CERS4
|
ceramide synthase 4 |
| chr12_+_77158021 | 0.12 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr2_+_26915584 | 0.12 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr16_-_46865286 | 0.12 |
ENST00000285697.4
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr14_+_56046914 | 0.12 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_-_150346607 | 0.12 |
ENST00000367341.1
ENST00000286380.2 |
RAET1L
|
retinoic acid early transcript 1L |
| chr7_-_44365020 | 0.12 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr12_-_106641728 | 0.12 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr6_+_143772060 | 0.12 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr7_-_27239703 | 0.12 |
ENST00000222753.4
|
HOXA13
|
homeobox A13 |
| chr17_-_57184170 | 0.12 |
ENST00000393065.2
|
TRIM37
|
tripartite motif containing 37 |
| chr6_+_12012170 | 0.12 |
ENST00000487103.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_+_41445413 | 0.12 |
ENST00000541520.1
|
CTPS1
|
CTP synthase 1 |
| chr8_-_94753184 | 0.12 |
ENST00000520560.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr17_+_5973793 | 0.12 |
ENST00000317744.5
|
WSCD1
|
WSC domain containing 1 |
| chr13_+_37393351 | 0.12 |
ENST00000255476.2
|
RFXAP
|
regulatory factor X-associated protein |
| chr15_+_79165296 | 0.12 |
ENST00000558746.1
ENST00000558830.1 ENST00000559345.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr18_+_48086440 | 0.12 |
ENST00000400384.2
ENST00000540640.1 ENST00000592595.1 |
MAPK4
|
mitogen-activated protein kinase 4 |
| chr13_+_76123883 | 0.12 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr2_+_181845532 | 0.11 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr3_+_51422478 | 0.11 |
ENST00000528157.1
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr8_+_29953163 | 0.11 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr14_-_105635090 | 0.11 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr15_+_79165372 | 0.11 |
ENST00000558502.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr2_-_136743436 | 0.11 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr9_-_3525968 | 0.11 |
ENST00000382004.3
ENST00000302303.1 ENST00000449190.1 |
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr10_-_105110890 | 0.11 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr17_+_7348658 | 0.11 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr9_-_15510989 | 0.11 |
ENST00000380715.1
ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr11_-_93474645 | 0.11 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr15_-_49912987 | 0.11 |
ENST00000560246.1
ENST00000558594.1 |
FAM227B
|
family with sequence similarity 227, member B |
| chr9_-_4741217 | 0.11 |
ENST00000447596.4
|
AK3
|
adenylate kinase 3 |
| chr17_+_75137460 | 0.11 |
ENST00000587820.1
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr1_+_236305826 | 0.11 |
ENST00000366592.3
ENST00000366591.4 |
GPR137B
|
G protein-coupled receptor 137B |
| chr15_+_79165222 | 0.11 |
ENST00000559930.1
|
MORF4L1
|
mortality factor 4 like 1 |
| chr2_-_73511559 | 0.11 |
ENST00000521871.1
|
FBXO41
|
F-box protein 41 |
| chr2_-_120124258 | 0.11 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr2_+_9563697 | 0.11 |
ENST00000238112.3
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr15_+_41136216 | 0.11 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr9_-_86571628 | 0.11 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr19_+_46144884 | 0.11 |
ENST00000593161.1
|
AC006132.1
|
chromosome 19 open reading frame 83 |
| chr1_-_45805752 | 0.10 |
ENST00000354383.6
ENST00000355498.2 ENST00000372100.5 ENST00000531105.1 |
MUTYH
|
mutY homolog |
| chr8_-_94753202 | 0.10 |
ENST00000521947.1
|
RBM12B
|
RNA binding motif protein 12B |
| chr2_+_149402009 | 0.10 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr19_+_17392672 | 0.10 |
ENST00000594072.1
ENST00000598347.1 |
ANKLE1
|
ankyrin repeat and LEM domain containing 1 |
| chr2_+_135676381 | 0.10 |
ENST00000537343.1
ENST00000295238.6 ENST00000264157.5 |
CCNT2
|
cyclin T2 |
| chr2_+_9564013 | 0.10 |
ENST00000460593.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr19_-_39832563 | 0.10 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr8_-_28243590 | 0.10 |
ENST00000523095.1
ENST00000522795.1 |
ZNF395
|
zinc finger protein 395 |
| chr1_-_63153944 | 0.10 |
ENST00000340370.5
ENST00000404627.2 ENST00000251157.5 |
DOCK7
|
dedicator of cytokinesis 7 |
| chr2_+_64681641 | 0.10 |
ENST00000409537.2
|
LGALSL
|
lectin, galactoside-binding-like |
| chr2_+_9563769 | 0.10 |
ENST00000475482.1
|
CPSF3
|
cleavage and polyadenylation specific factor 3, 73kDa |
| chr12_-_89920030 | 0.10 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chrX_+_21959108 | 0.10 |
ENST00000457085.1
|
SMS
|
spermine synthase |
| chr2_+_170655789 | 0.10 |
ENST00000409333.1
|
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr13_+_52586517 | 0.10 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr19_+_8274204 | 0.10 |
ENST00000561053.1
ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4
|
ceramide synthase 4 |
| chr22_+_20104947 | 0.10 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr20_+_31407692 | 0.10 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr5_-_6633397 | 0.10 |
ENST00000264670.6
ENST00000539938.1 |
NSUN2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr19_+_19496728 | 0.10 |
ENST00000537887.1
ENST00000417582.2 |
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr22_-_50964849 | 0.10 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr10_-_121302195 | 0.10 |
ENST00000369103.2
|
RGS10
|
regulator of G-protein signaling 10 |
| chr1_+_207494853 | 0.10 |
ENST00000367064.3
ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr2_+_54198210 | 0.10 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_120124383 | 0.10 |
ENST00000334816.7
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr3_-_148804275 | 0.10 |
ENST00000392912.2
ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF
|
helicase-like transcription factor |
| chr2_-_47142884 | 0.10 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr15_-_32162833 | 0.10 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr9_-_4741255 | 0.10 |
ENST00000381809.3
|
AK3
|
adenylate kinase 3 |
| chr3_+_134205000 | 0.10 |
ENST00000512894.1
ENST00000513612.2 ENST00000606977.1 |
CEP63
|
centrosomal protein 63kDa |
| chr4_+_57302297 | 0.10 |
ENST00000399688.3
ENST00000512576.1 |
PAICS
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr9_+_133971909 | 0.10 |
ENST00000247291.3
ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1-like |
| chr4_+_113152978 | 0.10 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr1_-_205290865 | 0.10 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr7_-_140341251 | 0.10 |
ENST00000491728.1
|
DENND2A
|
DENN/MADD domain containing 2A |
| chr8_+_56685701 | 0.09 |
ENST00000260129.5
|
TGS1
|
trimethylguanosine synthase 1 |
| chr20_+_61273797 | 0.09 |
ENST00000217159.1
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1 |
| chr10_+_16478942 | 0.09 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr15_-_44069741 | 0.09 |
ENST00000319359.3
|
ELL3
|
elongation factor RNA polymerase II-like 3 |
| chr8_-_66754172 | 0.09 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr6_+_151646800 | 0.09 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_-_113746277 | 0.09 |
ENST00000003302.4
ENST00000545540.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr9_-_124976185 | 0.09 |
ENST00000464484.2
|
LHX6
|
LIM homeobox 6 |
| chr12_+_122064673 | 0.09 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr9_-_86153218 | 0.09 |
ENST00000304195.3
ENST00000376438.1 |
FRMD3
|
FERM domain containing 3 |
| chr7_-_105925558 | 0.09 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr7_-_6098770 | 0.09 |
ENST00000536084.1
ENST00000446699.1 ENST00000199389.6 |
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr6_-_105850937 | 0.09 |
ENST00000369110.3
|
PREP
|
prolyl endopeptidase |
| chr6_+_86159765 | 0.09 |
ENST00000369646.3
ENST00000257770.3 |
NT5E
|
5'-nucleotidase, ecto (CD73) |
| chr3_-_185542761 | 0.09 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr9_+_33025209 | 0.09 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr1_-_8086343 | 0.09 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr12_-_107487604 | 0.09 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr2_+_29117509 | 0.09 |
ENST00000407426.3
|
WDR43
|
WD repeat domain 43 |
| chr13_-_51417812 | 0.09 |
ENST00000504404.1
|
DLEU7
|
deleted in lymphocytic leukemia, 7 |
| chr2_-_200715573 | 0.09 |
ENST00000420922.2
|
FTCDNL1
|
formiminotransferase cyclodeaminase N-terminal like |
| chr11_-_113746212 | 0.09 |
ENST00000537642.1
ENST00000537706.1 ENST00000544750.1 ENST00000260188.5 ENST00000540925.1 |
USP28
|
ubiquitin specific peptidase 28 |
| chr8_-_101348408 | 0.09 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr7_+_151038785 | 0.09 |
ENST00000413040.2
ENST00000568733.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr3_-_112738490 | 0.09 |
ENST00000393857.2
|
C3orf17
|
chromosome 3 open reading frame 17 |
| chr13_-_102068706 | 0.09 |
ENST00000251127.6
|
NALCN
|
sodium leak channel, non-selective |
| chr1_-_41707778 | 0.09 |
ENST00000337495.5
ENST00000372597.1 ENST00000372596.1 |
SCMH1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr14_+_93260569 | 0.09 |
ENST00000163416.2
|
GOLGA5
|
golgin A5 |
| chr3_-_185542817 | 0.09 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr15_+_73344911 | 0.09 |
ENST00000560262.1
ENST00000558964.1 |
NEO1
|
neogenin 1 |
| chr9_+_133971863 | 0.09 |
ENST00000372309.3
|
AIF1L
|
allograft inflammatory factor 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 0.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.1 | 0.2 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.3 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.4 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.0 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.0 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.2 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.0 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:2000619 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.0 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:2000323 | glucocorticoid secretion(GO:0035933) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) regulation of glucocorticoid secretion(GO:2000849) |
| 0.0 | 0.0 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.0 | GO:0002572 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.4 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.4 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0022840 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |