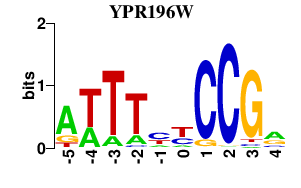
Results for YPR196W
Z-value: 0.72
Transcription factors associated with YPR196W
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
|
S000006400 | Putative maltose-responsive transcription factor |
Activity-expression correlation:
Activity profile of YPR196W motif
Sorted Z-values of YPR196W motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFR056C | 2.82 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YHR095W | 2.66 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR094C | 2.44 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YGL035C | 1.97 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YNL178W | 1.90 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YFR054C | 1.84 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR278C | 1.83 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKR075C | 1.80 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YPL026C | 1.68 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YGL033W | 1.65 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YFR055W | 1.49 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YJL106W | 1.46 |
IME2
|
Serine/threonine protein kinase involved in activation of meiosis, associates with Ime1p and mediates its stability, activates Ndt80p; IME2 expression is positively regulated by Ime1p |
|
| YJL214W | 1.46 |
HXT8
|
Protein of unknown function with similarity to hexose transporter family members, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YJL107C | 1.39 |
Putative protein of unknown function; expression is induced by activation of the HOG1 mitogen-activated signaling pathway and this induction is Hog1p/Pbs2p dependent; YJL107C and adjacent ORF, YJL108C are merged in related fungi |
||
| YNL179C | 1.39 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YGR140W | 1.35 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YLR308W | 1.29 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YDR043C | 1.28 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YNL180C | 1.19 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YNL065W | 1.14 |
AQR1
|
Plasma membrane multidrug transporter of the major facilitator superfamily, confers resistance to short-chain monocarboxylic acids and quinidine; involved in the excretion of excess amino acids |
|
| YAR069C | 1.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR093W | 1.04 |
AHT1
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; multicopy suppressor of glucose transport defects, likely due to the presence of an HXT4 regulatory element in the region |
|
| YLR154C | 1.03 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YGR097W | 1.02 |
ASK10
|
Component of the RNA polymerase II holoenzyme, phosphorylated in response to oxidative stress; has a role in destruction of Ssn8p, which relieves repression of stress-response genes |
|
| YJL215C | 1.02 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR047C | 1.02 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YGL209W | 0.99 |
MIG2
|
Protein containing zinc fingers, involved in repression, along with Mig1p, of SUC2 (invertase) expression by high levels of glucose; binds to Mig1p-binding sites in SUC2 promoter |
|
| YPR169W-A | 0.99 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YKL110C | 0.98 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YDL241W | 0.97 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YKL063C | 0.97 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YML088W | 0.95 |
UFO1
|
F-box receptor protein, subunit of the Skp1-Cdc53-F-box receptor (SCF) E3 ubiquitin ligase complex; binds to phosphorylated Ho endonuclease, allowing its ubiquitylation by SCF and subsequent degradation |
|
| YNR062C | 0.95 |
Putative membrane protein of unknown function |
||
| YKR099W | 0.95 |
BAS1
|
Myb-related transcription factor involved in regulating basal and induced expression of genes of the purine and histidine biosynthesis pathways; also involved in regulation of meiotic recombination at specific genes |
|
| YGL177W | 0.93 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL045W | 0.93 |
PSK2
|
One of two (see also PSK1) PAS domain containing S/T protein kinases; regulates sugar flux and translation in response to an unknown metabolite by phosphorylating Ugp1p and Gsy2p (sugar flux) and Caf20p, Tif11p and Sro9p (translation) |
|
| YOR306C | 0.92 |
MCH5
|
Plasma membrane riboflavin transporter; facilitates the uptake of vitamin B2; required for FAD-dependent processes; sequence similarity to mammalian monocarboxylate permeases, however mutants are not deficient in monocarboxylate transport |
|
| YOR030W | 0.92 |
DFG16
|
Probable multiple transmembrane protein, involved in diploid invasive and pseudohyphal growth upon nitrogen starvation; required for accumulation of processed Rim101p |
|
| YMR290W-A | 0.90 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YDR279W | 0.85 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YBL063W | 0.80 |
KIP1
|
Kinesin-related motor protein required for mitotic spindle assembly and chromosome segregation; functionally redundant with Cin8p |
|
| YOR096W | 0.80 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YGR242W | 0.79 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YAP1802/YGR241C |
||
| YDL211C | 0.79 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YLR346C | 0.77 |
Putative protein of unknown function found in mitochondria; expression is regulated by transcription factors involved in pleiotropic drug resistance, Pdr1p and Yrr1p; YLR346C is not an essential gene |
||
| YDR275W | 0.76 |
BSC2
|
Protein of unknown function, ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YLR124W | 0.75 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL062W | 0.74 |
MSN4
|
Transcriptional activator related to Msn2p; activated in stress conditions, which results in translocation from the cytoplasm to the nucleus; binds DNA at stress response elements of responsive genes, inducing gene expression |
|
| YLR108C | 0.67 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; YLR108C is not an esssential gene |
||
| YBR180W | 0.66 |
DTR1
|
Putative dityrosine transporter, required for spore wall synthesis; expressed during sporulation; member of the major facilitator superfamily (DHA1 family) of multidrug resistance transporters |
|
| YOL156W | 0.65 |
HXT11
|
Putative hexose transporter that is nearly identical to Hxt9p, has similarity to major facilitator superfamily (MFS) transporters and is involved in pleiotropic drug resistance |
|
| YAR070C | 0.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR178W | 0.62 |
STB5
|
Activator of multidrug resistance genes, forms a heterodimer with Pdr1p; contains a Zn(II)2Cys6 zinc finger domain that interacts with a PDRE (pleotropic drug resistance element) in vitro; binds Sin3p in a two-hybrid assay |
|
| YNR063W | 0.60 |
Putative zinc-cluster protein of unknown function |
||
| YDL007C-A | 0.60 |
Putative protein of unknown function |
||
| YJL162C | 0.60 |
JJJ2
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YNL235C | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF SIN4/YNL236W, a subunit of the mediator complex |
||
| YIL056W | 0.58 |
VHR1
|
Transcriptional activator, required for the vitamin H-responsive element (VHRE) mediated induction of VHT1 (Vitamin H transporter) and BIO5 (biotin biosynthesis intermediate transporter) in response to low biotin concentrations |
|
| YPL075W | 0.58 |
GCR1
|
Transcriptional activator of genes involved in glycolysis; DNA-binding protein that interacts and functions with the transcriptional activator Gcr2p |
|
| YGR034W | 0.58 |
RPL26B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Ap and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YJL105W | 0.58 |
SET4
|
Protein of unknown function, contains a SET domain |
|
| YMR017W | 0.57 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YOR050C | 0.57 |
Hypothetical protein |
||
| YGL180W | 0.56 |
ATG1
|
Protein serine/threonine kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway; structurally required for pre-autophagosome formation; during autophagy forms a complex with Atg13p and Atg17p |
|
| YDL022C-A | 0.56 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene DIA3; identified by fungal homology and RT-PCR |
||
| YKL109W | 0.56 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YKR034W | 0.55 |
DAL80
|
Negative regulator of genes in multiple nitrogen degradation pathways; expression is regulated by nitrogen levels and by Gln3p; member of the GATA-binding family, forms homodimers and heterodimers with Deh1p |
|
| YAL018C | 0.55 |
Putative protein of unknown function |
||
| YLR154W-C | 0.53 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YDR299W | 0.52 |
BFR2
|
Essential protein possibly involved in secretion; multicopy suppressor of sensitivity to Brefeldin A |
|
| YJL112W | 0.52 |
MDV1
|
Peripheral protein of the cytosolic face of the mitochondrial outer membrane, required for mitochondrial fission; interacts with Fis1p and with the dynamin-related GTPase Dnm1p; contains WD repeats |
|
| YOR063W | 0.52 |
RPL3
|
Protein component of the large (60S) ribosomal subunit, has similarity to E. coli L3 and rat L3 ribosomal proteins; involved in the replication and maintenance of killer double stranded RNA virus |
|
| YAR053W | 0.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL047W | 0.50 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YPR119W | 0.50 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YBL085W | 0.49 |
BOI1
|
Protein implicated in polar growth, functionally redundant with Boi2p; interacts with bud-emergence protein Bem1p; contains an SH3 (src homology 3) domain and a PH (pleckstrin homology) domain |
|
| YDL048C | 0.49 |
STP4
|
Protein containing a Kruppel-type zinc-finger domain; has similarity to Stp1p, Stp2p, and Stp3p |
|
| YBR292C | 0.49 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YOR029W | 0.48 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YLR013W | 0.48 |
GAT3
|
Protein containing GATA family zinc finger motifs |
|
| YLL012W | 0.48 |
YEH1
|
Steryl ester hydrolase, one of three gene products (Yeh1p, Yeh2p, Tgl1p) responsible for steryl ester hydrolase activity and involved in sterol homeostasis; localized to lipid particle membranes |
|
| YOL100W | 0.48 |
PKH2
|
Serine/threonine protein kinase involved in sphingolipid-mediated signaling pathway that controls endocytosis; activates Ypk1p and Ykr2p, components of signaling cascade required for maintenance of cell wall integrity; redundant with Pkh1p |
|
| YGL178W | 0.48 |
MPT5
|
Member of the Puf family of RNA-binding proteins; binds to mRNAs encoding chromatin modifiers and spindle pole body components; involved in longevity, maintenance of cell wall integrity, and sensitivity to and recovery from pheromone arrest |
|
| YKL221W | 0.47 |
MCH2
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YOR130C | 0.47 |
ORT1
|
Ornithine transporter of the mitochondrial inner membrane, exports ornithine from mitochondria as part of arginine biosynthesis; human ortholog is associated with hyperammonaemia-hyperornithinaemia-homocitrullinuria (HHH) syndrome |
|
| YKR039W | 0.47 |
GAP1
|
General amino acid permease; localization to the plasma membrane is regulated by nitrogen source |
|
| YPL141C | 0.47 |
Putative protein kinase; similar to Kin4p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YPL141C is not an essential gene |
||
| YGL158W | 0.45 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YBR081C | 0.45 |
SPT7
|
Subunit of the SAGA transcriptional regulatory complex, involved in proper assembly of the complex; also present as a C-terminally truncated form in the SLIK/SALSA transcriptional regulatory complex |
|
| YGL201C | 0.45 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YOR274W | 0.44 |
MOD5
|
Delta 2-isopentenyl pyrophosphate:tRNA isopentenyl transferase, required for biosynthesis of the modified base isopentenyladenosine in mitochondrial and cytoplasmic tRNAs; gene is nuclear and encodes two isozymic forms |
|
| YER169W | 0.44 |
RPH1
|
JmjC domain-containing histone demethylase which can specifically demethylate H3K36 tri- and dimethyl modification states; transcriptional repressor of PHR1; Rph1p phosphorylation during DNA damage is under control of the MEC1-RAD53 pathway |
|
| YBR034C | 0.44 |
HMT1
|
Nuclear SAM-dependent mono- and asymmetric arginine dimethylating methyltransferase that modifies hnRNPs, including Npl3p and Hrp1p, thus facilitating nuclear export of these proteins; required for viability of npl3 mutants |
|
| YPL025C | 0.44 |
Hypothetical protein |
||
| YOR388C | 0.44 |
FDH1
|
NAD(+)-dependent formate dehydrogenase, may protect cells from exogenous formate |
|
| YCR092C | 0.43 |
MSH3
|
Mismatch repair protein, forms dimers with Msh2p that mediate repair of insertion or deletion mutations and removal of nonhomologous DNA ends, contains a PCNA (Pol30p) binding motif required for genome stability |
|
| YGR241C | 0.43 |
YAP1802
|
Protein involved in clathrin cage assembly; binds Pan1p and clathrin; homologous to Yap1801p, member of the AP180 protein family |
|
| YPL062W | 0.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YOR062C | 0.41 |
Protein of unknown function; similar to YKR075Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YML053C | 0.41 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus; YML053C is not an essential gene |
||
| YDR259C | 0.41 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YDR274C | 0.40 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR008C-A | 0.40 |
Putative protein of unknown function, includes a potential transmembrane domain; deletion results in slightly lengthened telomeres |
||
| YNR072W | 0.40 |
HXT17
|
Hexose transporter, up-regulated in media containing raffinose and galactose at pH 7.7 versus pH 4.7, repressed by high levels of glucose |
|
| YIL118W | 0.40 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YMR037C | 0.40 |
MSN2
|
Transcriptional activator related to Msn4p; activated in stress conditions, which results in translocation from the cytoplasm to the nucleus; binds DNA at stress response elements of responsive genes, inducing gene expression |
|
| YGR287C | 0.40 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; has similarity to alpha-D-glucosidase (maltase); authentic, non-tagged protein detected in purified mitochondria in high-throughput studies |
||
| YPR171W | 0.40 |
BSP1
|
Adapter that links synaptojanins Inp52p and Inp53p to the cortical actin cytoskeleton |
|
| YMR290C | 0.39 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YKR076W | 0.39 |
ECM4
|
Omega class glutathione transferase; not essential; similar to Ygr154cp; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
|
| YDR145W | 0.39 |
TAF12
|
Subunit (61/68 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation and in chromatin modification, similar to histone H2A |
|
| YNR001W-A | 0.38 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YKL166C | 0.38 |
TPK3
|
cAMP-dependent protein kinase catalytic subunit; promotes vegetative growth in response to nutrients via the Ras-cAMP signaling pathway; inhibited by regulatory subunit Bcy1p in the absence of cAMP; partially redundant with Tpk1p and Tpk2p |
|
| YMR102C | 0.38 |
Protein of unknown function; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance; mutant shows increased resistance to azoles; YMR102C is not an essential gene |
||
| YFL015W-A | 0.38 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBL037W | 0.38 |
APL3
|
Alpha-adaptin, large subunit of the clathrin associated protein complex (AP-2); involved in vesicle mediated transport |
|
| YIL119C | 0.38 |
RPI1
|
Putative transcriptional regulator; overexpression suppresses the heat shock sensitivity of wild-type RAS2 overexpression and also suppresses the cell lysis defect of an mpk1 mutation |
|
| YIL057C | 0.38 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YEL007W | 0.37 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YGR251W | 0.37 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YOR101W | 0.37 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YGR051C | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YCR093W | 0.36 |
CDC39
|
Component of the CCR4-NOT complex, which has multiple roles in regulating mRNA levels including regulation of transcription and destabilizing mRNAs by deadenylation; basal transcription factor |
|
| YDR096W | 0.36 |
GIS1
|
JmjC domain-containing histone demethylase; transcription factor involved in the expression of genes during nutrient limitation; also involved in the negative regulation of DPP1 and PHR1 |
|
| YER137C | 0.36 |
Putative protein of unknown function |
||
| YBR114W | 0.36 |
RAD16
|
Protein that recognizes and binds damaged DNA in an ATP-dependent manner (with Rad7p) during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 4 (NEF4) and the Elongin-Cullin-Socs (ECS) ligase complex |
|
| YOR058C | 0.36 |
ASE1
|
Mitotic spindle midzone localized microtubule-associated protein (MAP) family member; required for spindle elongation and stabilization; undergoes cell cycle-regulated degradation by anaphase promoting complex; potential Cdc28p substrate |
|
| YMR124W | 0.36 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery, cytoplasm, bud, and bud neck; interacts with Crm1p in two-hybrid assay; YMR124W is not an essential gene |
||
| YBR238C | 0.36 |
Mitochondrial membrane protein with similarity to Rmd9p; not required for respiratory growth but causes a synthetic respiratory defect in combination with rmd9 mutations; transcriptionally up-regulated by TOR; deletion increases life span |
||
| YMR100W | 0.35 |
MUB1
|
Protein of unknown function, deletion causes multi-budding phenotype; has similarity to Aspergillus nidulans samB gene |
|
| YMR016C | 0.35 |
SOK2
|
Nuclear protein that plays a regulatory role in the cyclic AMP (cAMP)-dependent protein kinase (PKA) signal transduction pathway; negatively regulates pseudohyphal differentiation; homologous to several transcription factors |
|
| YDR095C | 0.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YML091C | 0.35 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YOR345C | 0.35 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YLR272C | 0.34 |
YCS4
|
Non-SMC subunit of the condensin complex (Smc2p-Smc4p-Ycs4p-Brn1p-Ycg1p); required for establishment and maintenance of chromosome condensation, chromosome segregation, chromatin binding of condensin and silencing at the mating type locus |
|
| YDR170C | 0.34 |
SEC7
|
Guanine nucleotide exchange factor (GEF) for ADP ribosylation factors involved in proliferation of the Golgi, intra-Golgi transport and ER-to-Golgi transport; found in the cytoplasm and on Golgi-associated coated vesicles |
|
| YPL183C | 0.33 |
RTT10
|
Cytoplasmic protein with a role in regulation of Ty1 transposition |
|
| YGR265W | 0.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF MES1/YGR264C, which encodes methionyl-tRNA synthetase |
||
| YOR355W | 0.33 |
GDS1
|
Protein of unknown function, required for growth on glycerol as a carbon source; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YBR112C | 0.33 |
CYC8
|
General transcriptional co-repressor, acts together with Tup1p; also acts as part of a transcriptional co-activator complex that recruits the SWI/SNF and SAGA complexes to promoters |
|
| YHR006W | 0.33 |
STP2
|
Transcription factor, activated by proteolytic processing in response to signals from the SPS sensor system for external amino acids; activates transcription of amino acid permease genes |
|
| YDL187C | 0.33 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YML077W | 0.32 |
BET5
|
Component of the TRAPP (transport protein particle) complex, which plays an essential role in the vesicular transport from endoplasmic reticulum to Golgi |
|
| YER131W | 0.32 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YER054C | 0.32 |
GIP2
|
Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism; contains a conserved motif (GVNK motif) that is also found in Gac1p, Pig1p, and Pig2p |
|
| YMR036C | 0.32 |
MIH1
|
Protein tyrosine phosphatase involved in cell cycle control; regulates the phosphorylation state of Cdc28p; homolog of S. pombe cdc25 |
|
| YER135C | 0.32 |
Dubious open reading frame unlikely to encode a protein; YER135C is not an essential gene |
||
| YGL024W | 0.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially/completely overlaps the verified ORF PGD1/YGL025C |
||
| YPL016W | 0.32 |
SWI1
|
Subunit of the SWI/SNF chromatin remodeling complex, which regulates transcription by remodeling chromosomes; required for transcription of many genes, including ADH1, ADH2, GAL1, HO, INO1 and SUC2; can become prion [SWI+] |
|
| YIR019C | 0.32 |
MUC1
|
GPI-anchored cell surface glycoprotein (flocculin) required for pseudohyphal formation, invasive growth, flocculation, and biofilms; transcriptionally regulated by the MAPK pathway (via Ste12p and Tec1p) and the cAMP pathway (via Flo8p) |
|
| YLR111W | 0.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR255C | 0.31 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR107W | 0.31 |
RGS2
|
Negative regulator of glucose-induced cAMP signaling; directly activates the GTPase activity of the heterotrimeric G protein alpha subunit Gpa2p |
|
| YNL118C | 0.31 |
DCP2
|
Catalytic subunit of the Dcp1p-Dcp2p decapping enzyme complex, which removes the 5' cap structure from mRNAs prior to their degradation; member of the Nudix hydrolase family |
|
| YJL194W | 0.31 |
CDC6
|
Essential ATP-binding protein required for DNA replication, component of the pre-replicative complex (pre-RC) which requires ORC to associate with chromatin and is in turn required for Mcm2-7p DNA association; homologous to S. pombe Cdc18p |
|
| YER164W | 0.31 |
CHD1
|
Nucleosome remodeling factor that functions in regulation of transcription elongation; contains a chromo domain, a helicase domain and a DNA-binding domain; component of both the SAGA and SLIK complexes |
|
| YJR160C | 0.31 |
MPH3
|
Alpha-glucoside permease, transports maltose, maltotriose, alpha-methylglucoside, and turanose; identical to Mph2p; encoded in a subtelomeric position in a region likely to have undergone duplication |
|
| YER047C | 0.31 |
SAP1
|
Putative ATPase of the AAA family, interacts with the Sin1p transcriptional repressor in the two-hybrid system |
|
| YLR263W | 0.30 |
RED1
|
Protein component of the axial elements of the synaptonemal complex, involved in chromosome segregation during the first meiotic division; interacts with Hop1p; required for wild-type levels of Mek1p kinase activity |
|
| YIR028W | 0.30 |
DAL4
|
Allantoin permease; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YER059W | 0.30 |
PCL6
|
Pho85p cyclin of the Pho80p subfamily; forms the major Glc8p kinase together with Pcl7p and Pho85p; involved in the control of glycogen storage by Pho85p; stabilized by Elongin C binding |
|
| YIL013C | 0.30 |
PDR11
|
ATP-binding cassette (ABC) transporter, multidrug transporter involved in multiple drug resistance; mediates sterol uptake when sterol biosynthesis is compromisedregulated by Pdr1p; required for anaerobic growth |
|
| YER064C | 0.30 |
Non-essential nuclear protein; null mutation has global effects on transcription |
||
| YBR106W | 0.30 |
PHO88
|
Probable membrane protein, involved in phosphate transport; pho88 pho86 double null mutant exhibits enhanced synthesis of repressible acid phosphatase at high inorganic phosphate concentrations |
|
| YAL003W | 0.29 |
EFB1
|
Translation elongation factor 1 beta; stimulates nucleotide exchange to regenerate EF-1 alpha-GTP for the next elongation cycle; part of the EF-1 complex, which facilitates binding of aminoacyl-tRNA to the ribosomal A site |
|
| YKR032W | 0.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER037W | 0.29 |
PHM8
|
Protein of unknown function, expression is induced by low phosphate levels and by inactivation of Pho85p |
|
| YHR212W-A | 0.29 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YMR011W | 0.29 |
HXT2
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YBR007C | 0.29 |
DSF2
|
Deletion suppressor of mpt5 mutation |
|
| YOL051W | 0.29 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YLR448W | 0.29 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YER062C | 0.29 |
HOR2
|
One of two redundant DL-glycerol-3-phosphatases (RHR2/GPP1 encodes the other) involved in glycerol biosynthesis; induced in response to hyperosmotic stress and oxidative stress, and during the diauxic transition |
|
| YHR021C | 0.28 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YKL064W | 0.28 |
MNR2
|
Putative magnesium transporter; has similarity to Alr1p and Alr2p, which mediate influx of Mg2+ and other divalent cations |
|
| YER097W | 0.28 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR092W | 0.28 |
BUD4
|
Protein involved in bud-site selection and required for axial budding pattern; localizes with septins to bud neck in mitosis and may constitute an axial landmark for next round of budding; potential Cdc28p substrate |
|
| YFL034C-A | 0.28 |
RPL22B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl22Ap and to rat L22 ribosomal protein |
|
| YDR028C | 0.28 |
REG1
|
Regulatory subunit of type 1 protein phosphatase Glc7p, involved in negative regulation of glucose-repressible genes |
|
| YPL143W | 0.28 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YLR431C | 0.28 |
ATG23
|
Peripheral membrane protein required for the cytoplasm-to-vacuole targeting (Cvt) pathway; cycles between the pre-autophagosome (PAS) and non-PAS locations; forms a complex with Atg9p and Atg27p |
|
| YDL023C | 0.28 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YGR264C | 0.28 |
MES1
|
Methionyl-tRNA synthetase, forms a complex with glutamyl-tRNA synthetase (Gus1p) and Arc1p, which increases the catalytic efficiency of both tRNA synthetases; also has a role in nuclear export of tRNAs |
|
| YDR029W | 0.28 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL273W | 0.28 |
TOF1
|
Subunit of a replication-pausing checkpoint complex (Tof1p-Mrc1p-Csm3p) that acts at the stalled replication fork to promote sister chromatid cohesion after DNA damage, facilitating gap repair of damaged DNA; interacts with the MCM helicase |
|
| YDR441C | 0.28 |
APT2
|
Apparent pseudogene, not transcribed or translated under normal conditions; encodes a protein with similarity to adenine phosphoribosyltransferase, but artificially expressed protein exhibits no enzymatic activity |
|
| YLR191W | 0.27 |
PEX13
|
Integral peroxisomal membrane required for the translocation of peroxisomal matrix proteins, interacts with the PTS1 signal recognition factor Pex5p and the PTS2 signal recognition factor Pex7p, forms a complex with Pex14p and Pex17p |
|
| YDR499W | 0.27 |
LCD1
|
Essential protein required for the DNA integrity checkpoint pathways; interacts physically with Mec1p; putative homolog of S. pombe Rad26 and human ATRIP |
|
| YMR136W | 0.27 |
GAT2
|
Protein containing GATA family zinc finger motifs; similar to Gln3p and Dal80p; expression repressed by leucine |
|
| YIL144W | 0.27 |
TID3
|
Component of the evolutionarily conserved kinetochore-associated Ndc80 complex (Ndc80p-Nuf2p-Spc24p-Spc25p); conserved coiled-coil protein involved in chromosome segregation, spindle checkpoint activity, kinetochore assembly and clustering |
|
| YEL061C | 0.27 |
CIN8
|
Kinesin motor protein involved in mitotic spindle assembly and chromosome segregation |
|
| YJR128W | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YMR135W-A | 0.26 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR300C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YBR301W; YBR300C is not an essential gene |
||
| YHR046C | 0.26 |
INM1
|
Inositol monophosphatase, involved in biosynthesis of inositol and in phosphoinositide second messenger signaling; INM1 expression increases in the presence of inositol and decreases upon exposure to antibipolar drugs lithium and valproate |
|
| YLR223C | 0.26 |
IFH1
|
Essential protein with a highly acidic N-terminal domain; IFH1 exhibits genetic interactions with FHL1, overexpression interferes with silencing at telomeres and HM loci; potential Cdc28p substrate |
|
| YDR442W | 0.26 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL122W | 0.26 |
UBP1
|
Ubiquitin-specific protease that removes ubiquitin from ubiquitinated proteins; cleaves at the C terminus of ubiquitin fusions irrespective of their size; capable of cleaving polyubiquitin chains |
|
| YNL221C | 0.26 |
POP1
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends; binds to the RPR1 RNA subunit in RNase P |
|
| YPL219W | 0.26 |
PCL8
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk) to phosphorylate and regulate glycogen synthase, also activates Pho85p for Glc8p phosphorylation |
Network of associatons between targets according to the STRING database.
First level regulatory network of YPR196W
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:1900436 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.5 | 1.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.5 | 1.4 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.4 | 1.3 | GO:0061422 | positive regulation of transcription from RNA polymerase II promoter in response to alkaline pH(GO:0061422) |
| 0.4 | 1.5 | GO:0000092 | mitotic anaphase B(GO:0000092) |
| 0.3 | 1.0 | GO:0046495 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.3 | 1.6 | GO:1901072 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.3 | 1.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 1.1 | GO:0071406 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.2 | 0.8 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.2 | 0.6 | GO:0072363 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.2 | 1.0 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 1.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.2 | 0.6 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 1.1 | GO:0032973 | amino acid export(GO:0032973) |
| 0.2 | 1.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 1.8 | GO:0015758 | glucose transport(GO:0015758) |
| 0.2 | 1.0 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) |
| 0.2 | 1.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.4 | GO:0018195 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.4 | GO:0035953 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.1 | 1.0 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.1 | 0.7 | GO:0007188 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.1 | 2.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.1 | GO:0006110 | regulation of glycolytic process(GO:0006110) |
| 0.1 | 0.8 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.3 | GO:0044109 | cellular alcohol catabolic process(GO:0044109) |
| 0.1 | 0.3 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 0.3 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.1 | 0.5 | GO:0032233 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.3 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.4 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.1 | 0.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.3 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.1 | 0.2 | GO:0071482 | cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.4 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.1 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.4 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 2.4 | GO:0051445 | regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.1 | GO:2000002 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 | 0.4 | GO:0000735 | removal of nonhomologous ends(GO:0000735) |
| 0.1 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.1 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.1 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 | 0.3 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.1 | 0.3 | GO:0000279 | mitotic M phase(GO:0000087) M phase(GO:0000279) |
| 0.1 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.1 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 0.3 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.1 | 0.7 | GO:0031070 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.1 | 0.1 | GO:0097306 | positive regulation of transcription from RNA polymerase II promoter in response to ethanol(GO:0061410) cellular response to ethanol(GO:0071361) cellular response to alcohol(GO:0097306) |
| 0.1 | 0.1 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.1 | 0.6 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.1 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) osmosensory signaling via phosphorelay pathway(GO:0007234) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 1.4 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.2 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.3 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.1 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.1 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.2 | GO:0015883 | FAD transport(GO:0015883) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.2 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.2 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.1 | 0.2 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.1 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.1 | 1.3 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 7.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.2 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.0 | 0.2 | GO:0072479 | cellular response to biotic stimulus(GO:0071216) response to cell cycle checkpoint signaling(GO:0072396) response to mitotic cell cycle checkpoint signaling(GO:0072414) response to spindle checkpoint signaling(GO:0072417) response to mitotic spindle checkpoint signaling(GO:0072476) response to mitotic cell cycle spindle assembly checkpoint signaling(GO:0072479) response to spindle assembly checkpoint signaling(GO:0072485) negative regulation of protein import into nucleus during spindle assembly checkpoint(GO:1901925) |
| 0.0 | 0.2 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.0 | 0.2 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.0 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.7 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0032186 | cellular bud neck septin ring organization(GO:0032186) |
| 0.0 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.2 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.0 | 0.2 | GO:0001015 | snoRNA transcription from an RNA polymerase II promoter(GO:0001015) |
| 0.0 | 0.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0046320 | regulation of fatty acid beta-oxidation(GO:0031998) positive regulation of fatty acid beta-oxidation(GO:0032000) regulation of fatty acid oxidation(GO:0046320) positive regulation of fatty acid oxidation(GO:0046321) regulation of lipid catabolic process(GO:0050994) positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 | 0.2 | GO:0007535 | donor selection(GO:0007535) |
| 0.0 | 0.1 | GO:1902750 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:0031135 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.1 | GO:0046136 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.0 | 0.0 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.0 | 0.5 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0070649 | parallel actin filament bundle assembly(GO:0030046) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.0 | 0.0 | GO:0010962 | regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0019218 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 0.3 | GO:0030473 | establishment of localization by movement along microtubule(GO:0010970) nuclear migration along microtubule(GO:0030473) organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.1 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.0 | 0.4 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.4 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.2 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.0 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:2001023 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.0 | 0.1 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0031032 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.8 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 1.2 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0000921 | septin ring assembly(GO:0000921) |
| 0.0 | 0.1 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.0 | 0.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0055070 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.0 | 0.4 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 0.0 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.7 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.8 | GO:0070591 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0042173 | regulation of sporulation resulting in formation of a cellular spore(GO:0042173) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.1 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.0 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.0 | 0.2 | GO:0042886 | amide transport(GO:0042886) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.0 | 0.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0034201 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.1 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.0 | 0.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.3 | GO:0009304 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0019932 | second-messenger-mediated signaling(GO:0019932) |
| 0.0 | 0.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.0 | GO:0031047 | gene silencing involved in chronological cell aging(GO:0010978) gene silencing by RNA(GO:0031047) |
| 0.0 | 0.1 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.0 | GO:0001315 | age-dependent response to reactive oxygen species(GO:0001315) |
| 0.0 | 0.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.4 | GO:0051302 | regulation of cell division(GO:0051302) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay(GO:0070481) |
| 0.0 | 0.1 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.0 | 0.0 | GO:0090342 | regulation of cell aging(GO:0090342) regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0070478 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay(GO:0070478) |
| 0.0 | 0.1 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0007532 | regulation of mating-type specific transcription, DNA-templated(GO:0007532) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.0 | GO:0090522 | vesicle tethering involved in exocytosis(GO:0090522) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.8 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.0 | GO:0032262 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 0.0 | GO:0000433 | negative regulation of transcription from RNA polymerase II promoter by glucose(GO:0000433) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 2.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.3 | 1.4 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.2 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 0.9 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.2 | 1.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.1 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.1 | 0.3 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.1 | 1.0 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.1 | 0.5 | GO:0032545 | CURI complex(GO:0032545) |
| 0.1 | 0.4 | GO:0030669 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.2 | GO:0032177 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.1 | 1.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.4 | GO:0000796 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.1 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0070775 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 4.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.3 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.1 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.4 | GO:0005628 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.2 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0030880 | DNA-directed RNA polymerase complex(GO:0000428) RNA polymerase complex(GO:0030880) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0008278 | nuclear cohesin complex(GO:0000798) cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 1.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0000144 | cellular bud neck septin ring(GO:0000144) |
| 0.0 | 0.4 | GO:0000142 | cellular bud neck contractile ring(GO:0000142) |
| 0.0 | 0.1 | GO:0097344 | Rix1 complex(GO:0097344) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.5 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.1 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.0 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.1 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 0.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.0 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 1.6 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.0 | 0.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.5 | GO:0005935 | cellular bud neck(GO:0005935) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0098552 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.3 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0016587 | Isw1 complex(GO:0016587) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0000136 | alpha-1,6-mannosyltransferase complex(GO:0000136) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.0 | GO:0070823 | HDA1 complex(GO:0070823) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0035361 | Cul8-RING ubiquitin ligase complex(GO:0035361) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 12.8 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.5 | 1.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 2.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.3 | 5.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 0.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.2 | 1.4 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.4 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.2 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.2 | 0.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.4 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.3 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 1.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.1 | 0.5 | GO:0042887 | amide transmembrane transporter activity(GO:0042887) |
| 0.1 | 0.5 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 1.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.3 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.4 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.1 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.2 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.1 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.3 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.1 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.2 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.1 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 5.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 2.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 3.0 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 1.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 1.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 7.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0005186 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.0 | 0.1 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.0 | 1.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0090484 | drug transporter activity(GO:0090484) |
| 0.0 | 0.1 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.0 | 0.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.3 | GO:0019001 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.0 | 0.3 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.0 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 0.1 | GO:0008251 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.1 | GO:0070717 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.0 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.9 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.0 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 1.0 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.3 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0034212 | peptide alpha-N-acetyltransferase activity(GO:0004596) peptide N-acetyltransferase activity(GO:0034212) |
| 0.0 | 0.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.0 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 18.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 2.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 0.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.3 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.6 | REACTOME RNA POL II TRANSCRIPTION | Genes involved in RNA Polymerase II Transcription |
| 0.0 | 0.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.0 | 0.1 | REACTOME SIGNALLING TO ERKS | Genes involved in Signalling to ERKs |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 18.6 | REACTOME GLYCOSAMINOGLYCAN METABOLISM | Genes involved in Glycosaminoglycan metabolism |
| 0.0 | 0.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.0 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |