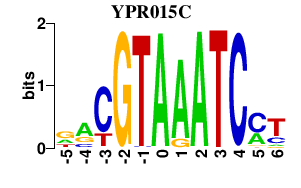
Results for YPR015C
Z-value: 0.29
Transcription factors associated with YPR015C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
|
S000006219 | Putative protein of unknown function |
Activity-expression correlation:
Activity profile of YPR015C motif
Sorted Z-values of YPR015C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YDR044W | 0.62 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YLR154W-A | 0.52 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-B | 0.48 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YDR033W | 0.48 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YNR001W-A | 0.48 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YBR158W | 0.44 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YOR107W | 0.43 |
RGS2
|
Negative regulator of glucose-induced cAMP signaling; directly activates the GTPase activity of the heterotrimeric G protein alpha subunit Gpa2p |
|
| YCL049C | 0.40 |
Protein of unknown function; localizes to membrane fraction; YCL049C is not an essential gene; |
||
| YIR021W | 0.38 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YLR314C | 0.36 |
CDC3
|
Component of the septin ring of the mother-bud neck that is required for cytokinesis; septins recruit proteins to the neck and can act as a barrier to diffusion at the membrane, and they comprise the 10nm filaments seen with EM |
|
| YPR157W | 0.36 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YMR083W | 0.36 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YIL009W | 0.32 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YKL164C | 0.32 |
PIR1
|
O-glycosylated protein required for cell wall stability; attached to the cell wall via beta-1,3-glucan; mediates mitochondrial translocation of Apn1p; expression regulated by the cell integrity pathway and by Swi5p during the cell cycle |
|
| YNL066W | 0.31 |
SUN4
|
Cell wall protein related to glucanases, possibly involved in cell wall septation; member of the SUN family |
|
| YLR413W | 0.31 |
Putative protein of unknown function; YLR413W is not an essential gene |
||
| YLR312W-A | 0.30 |
MRPL15
|
Mitochondrial ribosomal protein of the large subunit |
|
| YLR206W | 0.29 |
ENT2
|
Epsin-like protein required for endocytosis and actin patch assembly and functionally redundant with Ent1p; contains clathrin-binding motif at C-terminus |
|
| YMR106C | 0.29 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YIL009C-A | 0.28 |
EST3
|
Component of the telomerase holoenzyme, involved in telomere replication |
|
| YGL097W | 0.28 |
SRM1
|
Nucleotide exchange factor for Gsp1p, localizes to the nucleus, required for nucleocytoplasmic trafficking of macromolecules; suppressor of the pheromone response pathway; potentially phosphorylated by Cdc28p |
|
| YMR266W | 0.25 |
RSN1
|
Membrane protein of unknown function; overexpression suppresses NaCl sensitivity of sro7 mutant |
|
| YJL048C | 0.24 |
UBX6
|
UBX (ubiquitin regulatory X) domain-containing protein that interacts with Cdc48p, transcription is repressed when cells are grown in media containing inositol and choline |
|
| YDL055C | 0.24 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YBR177C | 0.23 |
EHT1
|
Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane |
|
| YDR098C | 0.22 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YGR086C | 0.22 |
PIL1
|
Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways; detected in phosphorylated state in mitochondria |
|
| YAL022C | 0.22 |
FUN26
|
Nucleoside transporter with broad nucleoside selectivity; localized to intracellular membranes |
|
| YNL234W | 0.21 |
Similar to globins and has a functional heme-binding domain; involved in glucose signaling or metabolism; regulated by Rgt1p |
||
| YER107C | 0.21 |
GLE2
|
Component of the nuclear pore complex required for polyadenylated RNA export but not for protein import, homologous to S. pombe Rae1p |
|
| YIL118W | 0.21 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YER107W-A | 0.21 |
Dubious open reading frame unlikely to encode a protein, partially overlaps verified ORF GLE2/YER107C |
||
| YCL024W | 0.21 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YDR100W | 0.21 |
TVP15
|
Integral membrane protein localized to late Golgi vesicles along with the v-SNARE Tlg2p |
|
| YGL009C | 0.20 |
LEU1
|
Isopropylmalate isomerase, catalyzes the second step in the leucine biosynthesis pathway |
|
| YGL012W | 0.20 |
ERG4
|
C-24(28) sterol reductase, catalyzes the final step in ergosterol biosynthesis; mutants are viable, but lack ergosterol |
|
| YDR297W | 0.20 |
SUR2
|
Sphinganine C4-hydroxylase, catalyses the conversion of sphinganine to phytosphingosine in sphingolipid biosyntheis |
|
| YDR185C | 0.20 |
Mitochondrial protein of unknown function; has similarity to Ups1p, which is involved in regulation of alternative topogenesis of the dynamin-related GTPase Mgm1p |
||
| YGL201C | 0.20 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YOL124C | 0.20 |
TRM11
|
Catalytic subunit of an adoMet-dependent tRNA methyltransferase complex (Trm11p-Trm112p), required for the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs; contains a THUMP domain and a methyltransferase domain |
|
| YMR265C | 0.19 |
Putative protein of unknown function |
||
| YER137C | 0.19 |
Putative protein of unknown function |
||
| YLR400W | 0.19 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR432W | 0.19 |
IMD3
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YMR043W | 0.19 |
MCM1
|
Transcription factor involved in cell-type-specific transcription and pheromone response; plays a central role in the formation of both repressor and activator complexes |
|
| YMR202W | 0.19 |
ERG2
|
C-8 sterol isomerase, catalyzes the isomerization of the delta-8 double bond to the delta-7 position at an intermediate step in ergosterol biosynthesis |
|
| YCL023C | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF KCC4 |
||
| YJL035C | 0.19 |
TAD2
|
Subunit of tRNA-specific adenosine-34 deaminase, forms a heterodimer with Tad3p that converts adenosine to inosine at the wobble position of several tRNAs |
|
| YLR154C | 0.19 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YLR154W-C | 0.18 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YMR320W | 0.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YFR055W | 0.18 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YKL051W | 0.18 |
SFK1
|
Plasma membrane protein that may act together with or upstream of Stt4p to generate normal levels of the essential phospholipid PI4P, at least partially mediates proper localization of Stt4p to the plasma membrane |
|
| YGR242W | 0.17 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YAP1802/YGR241C |
||
| YKL052C | 0.17 |
ASK1
|
Essential subunit of the Dam1 complex (aka DASH complex), couples kinetochores to the force produced by MT depolymerization thereby aiding in chromosome segregation; phosphorylated during the cell cycle by cyclin-dependent kinases |
|
| YHR183W | 0.17 |
GND1
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone and adaptation to oxidative stress |
|
| YOR099W | 0.17 |
KTR1
|
Alpha-1,2-mannosyltransferase involved in O- and N-linked protein glycosylation; type II membrane protein; member of the KRE2/MNT1 mannosyltransferase family |
|
| YNL087W | 0.17 |
TCB2
|
Bud-specific protein with a potential role in membrane trafficking; GFP-fusion protein migrates from the cell surface to intracellular vesicles near vacuole; contains 3 calcium and lipid binding domains; mRNA is targeted to the bud |
|
| YKL080W | 0.17 |
VMA5
|
Subunit C of the eight-subunit V1 peripheral membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane |
|
| YPR156C | 0.17 |
TPO3
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YBR017C | 0.16 |
KAP104
|
Transportin, cytosolic karyopherin beta 2 involved in delivery of heterogeneous nuclear ribonucleoproteins to the nucleoplasm, binds rg-nuclear localization signals on Nab2p and Hrp1p, plays a role in cell-cycle progression |
|
| YER131W | 0.16 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YKL096W-A | 0.16 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YPR074C | 0.16 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YHR181W | 0.16 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YOR047C | 0.16 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YER133W-A | 0.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps uncharacterized gene YER134C. |
||
| YEL068C | 0.16 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YKR043C | 0.16 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YML077W | 0.16 |
BET5
|
Component of the TRAPP (transport protein particle) complex, which plays an essential role in the vesicular transport from endoplasmic reticulum to Golgi |
|
| YPL172C | 0.15 |
COX10
|
Heme A:farnesyltransferase, catalyzes the first step in the conversion of protoheme to the heme A prosthetic group required for cytochrome c oxidase activity; human ortholog is associated with mitochondrial disorders |
|
| YPL004C | 0.15 |
LSP1
|
Primary component of eisosomes, which are large immobile patch structures at the cell cortex associated with endocytosis, along with Pil1p and Sur7p; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways |
|
| YER012W | 0.15 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YGR241C | 0.14 |
YAP1802
|
Protein involved in clathrin cage assembly; binds Pan1p and clathrin; homologous to Yap1801p, member of the AP180 protein family |
|
| YPR126C | 0.14 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL012C | 0.14 |
HTZ1
|
Histone variant H2AZ, exchanged for histone H2A in nucleosomes by the SWR1 complex; involved in transcriptional regulation through prevention of the spread of silent heterochromatin |
|
| YFR056C | 0.14 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YDL120W | 0.14 |
YFH1
|
Frataxin, regulates mitochondrial iron accumulation; interacts with Isu1p which promotes Fe-S cluster assembly; interacts with electron transport chain components and may influence respiration; human homolog involved in Friedrich's ataxia |
|
| YPL256C | 0.14 |
CLN2
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YDR084C | 0.14 |
TVP23
|
Integral membrane protein localized to late Golgi vesicles along with the v-SNARE Tlg2p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YMR173W | 0.14 |
DDR48
|
DNA damage-responsive protein, expression is increased in response to heat-shock stress or treatments that produce DNA lesions; contains multiple repeats of the amino acid sequence NNNDSYGS |
|
| YBR210W | 0.14 |
ERV15
|
Protein involved in export of proteins from the endoplasmic reticulum, has similarity to Erv14p |
|
| YFL015C | 0.14 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YGR249W | 0.14 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YPL003W | 0.13 |
ULA1
|
Protein that acts together with Uba3p to activate Rub1p before its conjugation to proteins (neddylation), which may play a role in protein degradation |
|
| YDL121C | 0.13 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic retiuculum; YDL121C is not an essential protein |
||
| YLR229C | 0.13 |
CDC42
|
Small rho-like GTPase, essential for establishment and maintenance of cell polarity; mutants have defects in the organization of actin and septins |
|
| YGR259C | 0.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF TNA1/YGR260W |
||
| YHR182C-A | 0.13 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR173W-A | 0.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene DDR48/YML173W |
||
| YEL066W | 0.13 |
HPA3
|
D-Amino acid N-acetyltransferase, catalyzes N-acetylation of D-amino acids through ordered bi-bi mechanism in which acetyl-CoA is first substrate bound and CoA is last product liberated; similar to Hpa2p, acetylates histones weakly in vitro |
|
| YLR029C | 0.13 |
RPL15A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Bp and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YKL097C | 0.13 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YLR333C | 0.13 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YMR199W | 0.13 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YGR036C | 0.13 |
CAX4
|
Dolichyl pyrophosphate (Dol-P-P) phosphatase with a luminally oriented active site in the ER, cleaves the anhydride linkage in Dol-P-P, required for Dol-P-P-linked oligosaccharide intermediate synthesis and protein N-glycosylation |
|
| YER134C | 0.12 |
Putative protein of unknown function; non-essential gene |
||
| YNL241C | 0.12 |
ZWF1
|
Glucose-6-phosphate dehydrogenase (G6PD), catalyzes the first step of the pentose phosphate pathway; involved in adapting to oxidatve stress; homolog of the human G6PD which is deficient in patients with hemolytic anemia |
|
| YJL173C | 0.12 |
RFA3
|
Subunit of heterotrimeric Replication Protein A (RPA), which is a highly conserved single-stranded DNA binding protein involved in DNA replication, repair, and recombination |
|
| YLR328W | 0.12 |
NMA1
|
Nicotinic acid mononucleotide adenylyltransferase, involved in pathways of NAD biosynthesis, including the de novo, NAD(+) salvage, and nicotinamide riboside salvage pathways |
|
| YER130C | 0.12 |
Hypothetical protein |
||
| YFL015W-A | 0.12 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR072C | 0.12 |
IPT1
|
Inositolphosphotransferase 1, involved in synthesis of mannose-(inositol-P)2-ceramide (M(IP)2C), which is the most abundant sphingolipid in cells, mutation confers resistance to the antifungals syringomycin E and DmAMP1 in some growth media |
|
| YKL218C | 0.12 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YFR042W | 0.12 |
KEG1
|
Integral membrane protein of the ER; physically interacts with Kre6p; has a role in the synthesis of beta-1,6-glucan in the cell wall; required for cell viability |
|
| YPR017C | 0.11 |
DSS4
|
Guanine nucleotide dissociation stimulator for Sec4p, functions in the post-Golgi secretory pathway; binds zinc, found both on membranes and in the cytosol |
|
| YBR028C | 0.11 |
Putative protein kinase, possible substrate of cAMP-dependent protein kinase (PKA) |
||
| YPR118W | 0.11 |
MRI1
|
Methylthioribose-1-phosphate isomerase, catalyzes the isomerization of 5-methylthioribose-1-phosphate to 5-methylthioribulose-1-phosphate in the methionine salvage pathway |
|
| YEL058W | 0.11 |
PCM1
|
Essential N-acetylglucosamine-phosphate mutase; converts GlcNAc-6-P to GlcNAc-1-P, which is a precursor for the biosynthesis of chitin and for the formation of N-glycosylated mannoproteins and glycosylphosphatidylinositol anchors |
|
| YPL178W | 0.11 |
CBC2
|
Small subunit of the heterodimeric cap binding complex that also contains Sto1p, component of the spliceosomal commitment complex; interacts with Npl3p, possibly to package mRNA for export from the nucleus; contains an RNA-binding motif |
|
| YOL011W | 0.11 |
PLB3
|
Phospholipase B (lysophospholipase) involved in phospholipid metabolism; hydrolyzes phosphatidylinositol and phosphatidylserine and displays transacylase activity in vitro |
|
| YGL022W | 0.11 |
STT3
|
Subunit of the oligosaccharyltransferase complex of the ER lumen, which catalyzes asparagine-linked glycosylation of newly synthesized proteins; forms a subcomplex with Ost3p and Ost4p and is directly involved in catalysis |
|
| YLR230W | 0.11 |
Dubious open reading frame unlikely to encode a functional protein; overlaps 5' end of essential CDC42 gene which encodes a small Rho-like GTPase essential for establishment and maintenance of cell polarity |
||
| YNR016C | 0.11 |
ACC1
|
Acetyl-CoA carboxylase, biotin containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA; required for de novo biosynthesis of long-chain fatty acids |
|
| YOR273C | 0.11 |
TPO4
|
Polyamine transport protein, recognizes spermine, putrescine, and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YDR133C | 0.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YDR348C | 0.11 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and bud neck; potential Cdc28p substrate |
||
| YNR017W | 0.11 |
TIM23
|
Essential protein of the mitochondrial inner membrane, component of the mitochondrial import system |
|
| YGR040W | 0.10 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YML054C-A | 0.10 |
Putative protein of unknown function |
||
| YDL171C | 0.10 |
GLT1
|
NAD(+)-dependent glutamate synthase (GOGAT), synthesizes glutamate from glutamine and alpha-ketoglutarate; with Gln1p, forms the secondary pathway for glutamate biosynthesis from ammonia; expression regulated by nitrogen source |
|
| YDR152W | 0.10 |
GIR2
|
Highly-acidic cytoplasmic RWD domain-containing protein of unknown function, sensitive to proteolysis, N-terminal region has high content of acidic amino acid residues, putative IUP (intrinsically unstructured protein) |
|
| YGL209W | 0.10 |
MIG2
|
Protein containing zinc fingers, involved in repression, along with Mig1p, of SUC2 (invertase) expression by high levels of glucose; binds to Mig1p-binding sites in SUC2 promoter |
|
| YOL101C | 0.10 |
IZH4
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by fatty acids and altered zinc levels; deletion reduces sensitivity to excess zinc; possible role in sterol metabolism |
|
| YMR172C-A | 0.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR080W | 0.10 |
TEF1
|
Translational elongation factor EF-1 alpha; also encoded by TEF2; functions in the binding reaction of aminoacyl-tRNA (AA-tRNA) to ribosomes |
|
| YBR247C | 0.10 |
ENP1
|
Protein associated with U3 and U14 snoRNAs, required for pre-rRNA processing and 40S ribosomal subunit synthesis; localized in the nucleus and concentrated in the nucleolus |
|
| YHR103W | 0.10 |
SBE22
|
Protein involved in the transport of cell wall components from the Golgi to the cell surface; similar in structure and functionally redundant with Sbe2p; involved in bud growth |
|
| YOR013W | 0.10 |
IRC11
|
Dubious opening reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YOR012C; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YNL169C | 0.09 |
PSD1
|
Phosphatidylserine decarboxylase of the mitochondrial inner membrane, converts phosphatidylserine to phosphatidylethanolamine |
|
| YGL020C | 0.09 |
GET1
|
Subunit of the GET complex; required for the retrieval of HDEL proteins from the Golgi to the ER in an ERD2 dependent fashion and for normal mitochondrial morphology and inheritance |
|
| YEL052W | 0.09 |
AFG1
|
Conserved protein that may act as a chaperone in the degradation of misfolded or unassembled cytochrome c oxidase subunits; localized to matrix face of the mitochondrial inner membrane; member of the AAA family but lacks a protease domain |
|
| YPL154C | 0.09 |
PEP4
|
Vacuolar aspartyl protease (proteinase A), required for the posttranslational precursor maturation of vacuolar proteinases; important for protein turnover after oxidative damage; synthesized as a zymogen, self-activates |
|
| YDR023W | 0.09 |
SES1
|
Cytosolic seryl-tRNA synthetase, class II aminoacyl-tRNA synthetase that aminoacylates tRNA(Ser), displays tRNA-dependent amino acid recognition which enhances discrimination of the serine substrate, interacts with peroxin Pex21p |
|
| YBR048W | 0.09 |
RPS11B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Ap and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YJR123W | 0.09 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YBR178W | 0.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YBR177C |
||
| YNL178W | 0.09 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YJR124C | 0.09 |
Putative protein of unknown function; expression induced under calcium shortage |
||
| YGR027C | 0.09 |
RPS25A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Bp and has similarity to rat S25 ribosomal protein |
|
| YNR003C | 0.09 |
RPC34
|
RNA polymerase III subunit C34; interacts with TFIIIB70 and is a key determinant in pol III recruitment by the preinitiation complex |
|
| YNL043C | 0.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YIP3/YNL044W |
||
| YMR082C | 0.09 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL058W | 0.09 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHL003C | 0.09 |
LAG1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lac1p |
|
| YOR155C | 0.09 |
ISN1
|
Inosine 5'-monophosphate (IMP)-specific 5'-nucleotidase, catalyzes the breakdown of IMP to inosine, does not show similarity to known 5'-nucleotidases from other organisms |
|
| YNL104C | 0.09 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YPR113W | 0.08 |
PIS1
|
Phosphatidylinositol synthase, required for biosynthesis of phosphatidylinositol, which is a precursor for polyphosphoinositides, sphingolipids, and glycolipid anchors for some of the plasma membrane proteins |
|
| YDR073W | 0.08 |
SNF11
|
Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; interacts with a highly conserved 40-residue sequence of Snf2p |
|
| YDR002W | 0.08 |
YRB1
|
Ran GTPase binding protein; involved in nuclear protein import and RNA export, ubiquitin-mediated protein degradation during the cell cycle; shuttles between the nucleus and cytoplasm; is essential; homolog of human RanBP1 |
|
| YGL019W | 0.08 |
CKB1
|
Beta regulatory subunit of casein kinase 2, a Ser/Thr protein kinase with roles in cell growth and proliferation; the holoenzyme also contains CKA1, CKA2 and CKB2, the many substrates include transcription factors and all RNA polymerases |
|
| YEL059C-A | 0.08 |
SOM1
|
Subunit of the mitochondrial inner membrane peptidase, which is required for maturation of mitochondrial proteins of the intermembrane space; Som1p facilitates cleavage of a subset of substrates |
|
| YLR120C | 0.08 |
YPS1
|
Aspartic protease, attached to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor |
|
| YLR353W | 0.08 |
BUD8
|
Protein involved in bud-site selection; diploid mutants display a unipolar budding pattern instead of the wild-type bipolar pattern, and bud at the proximal pole |
|
| YDR531W | 0.08 |
Pantothenate kinase (ATP:D-pantothenate 4'-phosphotransferase, EC 2.7.1.33); catalyzes the first committed step in the universal biosynthetic pathway leading to coenzyme A1 |
||
| YEL053C | 0.08 |
MAK10
|
Non-catalytic subunit of N-terminal acetyltransferase of the NatC type, required for replication of dsRNA virus; expression is glucose-repressible |
|
| YOL056W | 0.08 |
GPM3
|
Homolog of Gpm1p phosphoglycerate mutase, which converts 3-phosphoglycerate to 2-phosphoglycerate in glycolysis; may be non-functional derivative of a gene duplication event |
|
| YLR177W | 0.08 |
Putative protein of unknown function; phosphorylated by Dbf2p-Mob1p in vitro; some strains contain microsatellite polymophisms at this locus; YLR177W is not an essential gene |
||
| YJR001W | 0.08 |
AVT1
|
Vacuolar transporter, imports large neutral amino acids into the vacuole; member of a family of seven S. cerevisiae genes (AVT1-7) related to vesicular GABA-glycine transporters |
|
| YOR048C | 0.08 |
RAT1
|
Nuclear 5' to 3' single-stranded RNA exonuclease, involved in RNA metabolism, including rRNA and snRNA processing as well as mRNA transcription termination |
|
| YKL122C | 0.08 |
SRP21
|
Subunit of the signal recognition particle (SRP), which functions in protein targeting to the endoplasmic reticulum membrane; not found in mammalian SRP; forms a pre-SRP structure in the nucleolus that is translocated to the cytoplasm |
|
| YER147C-A | 0.08 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL272C | 0.08 |
Putative protein of unknown function; gene expression induced in response to ketoconazole; YPL272C is not an essential gene |
||
| YLR411W | 0.08 |
CTR3
|
High-affinity copper transporter of the plasma membrane, acts as a trimer; gene is disrupted by a Ty2 transposon insertion in many laboratory strains of S. cerevisiae |
|
| YNL289W | 0.08 |
PCL1
|
Pho85 cyclin of the Pcl1,2-like subfamily, involved in entry into the mitotic cell cycle and regulation of morphogenesis, localizes to sites of polarized cell growth |
|
| YBR032W | 0.08 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YDL084W | 0.08 |
SUB2
|
Component of the TREX complex required for nuclear mRNA export; member of the DEAD-box RNA helicase superfamily and is involved in early and late steps of spliceosome assembly; homolog of the human splicing factor hUAP56 |
|
| YPL274W | 0.08 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YLR257W | 0.08 |
Putative protein of unknown function |
||
| YKL110C | 0.07 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YKR013W | 0.07 |
PRY2
|
Protein of unknown function, has similarity to Pry1p and Pry3p and to the plant PR-1 class of pathogen related proteins |
|
| YDL211C | 0.07 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YGR265W | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF MES1/YGR264C, which encodes methionyl-tRNA synthetase |
||
| YFR041C | 0.07 |
ERJ5
|
Type I membrane protein with a J domain is required to preserve the folding capacity of the endoplasmic reticulum; loss of the non-essential ERJ5 gene leads to a constitutively induced unfolded protein response |
|
| YDR237W | 0.07 |
MRPL7
|
Mitochondrial ribosomal protein of the large subunit |
|
| YPR170W-B | 0.07 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YMR009W | 0.07 |
ADI1
|
Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions |
|
| YMR208W | 0.07 |
ERG12
|
Mevalonate kinase, acts in the biosynthesis of isoprenoids and sterols, including ergosterol, from mevalonate |
|
| YCR028C-A | 0.07 |
RIM1
|
Single-stranded DNA-binding protein essential for mitochondrial genome maintenance; involved in mitochondrial DNA replication |
|
| YDR375C | 0.07 |
BCS1
|
Protein of the mitochondrial inner membrane that functions as an ATP-dependent chaperone, required for the incorporation of the Rip1p and Qcr10p subunits into the cytochrome bc(1) complex; member of the CDC48/PAS1/SEC18 ATPase family |
|
| YMR011W | 0.07 |
HXT2
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YGR177C | 0.07 |
ATF2
|
Alcohol acetyltransferase, may play a role in steroid detoxification; forms volatile esters during fermentation, which is important in brewing |
|
| YHR180W-A | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YLL027W | 0.06 |
ISA1
|
Mitochondrial matrix protein involved in biogenesis of the iron-sulfur (Fe/S) cluster of Fe/S proteins, isa1 deletion causes loss of mitochondrial DNA and respiratory deficiency; depletion reduces growth on nonfermentable carbon sources |
|
| YML006C | 0.06 |
GIS4
|
CAAX box containing protein of unknown function, proposed to be involved in the RAS/cAMP signaling pathway |
|
| YLR002C | 0.06 |
NOC3
|
Protein that forms a nuclear complex with Noc2p that binds to 66S ribosomal precursors to mediate their intranuclear transport; also binds to chromatin to promote the association of DNA replication factors and replication initiation |
|
| YAL007C | 0.06 |
ERP2
|
Protein that forms a heterotrimeric complex with Erp1p, Emp24p, and Erv25p; member, along with Emp24p and Erv25p, of the p24 family involved in ER to Golgi transport and localized to COPII-coated vesicles |
|
| YFR035C | 0.06 |
Putative protein of unknown function, deletion mutant exhibits synthetic phenotype with alpha-synuclein |
||
| YDR261C | 0.06 |
EXG2
|
Exo-1,3-beta-glucanase, involved in cell wall beta-glucan assembly; may be anchored to the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor |
|
| YOR043W | 0.06 |
WHI2
|
Protein required, with binding partner Psr1p, for full activation of the general stress response, possibly through Msn2p dephosphorylation; regulates growth during the diauxic shift; negative regulator of G1 cyclin expression |
|
| YNL173C | 0.06 |
MDG1
|
Plasma membrane protein involved in G-protein mediated pheromone signaling pathway; overproduction suppresses bem1 mutations |
|
| YGR264C | 0.06 |
MES1
|
Methionyl-tRNA synthetase, forms a complex with glutamyl-tRNA synthetase (Gus1p) and Arc1p, which increases the catalytic efficiency of both tRNA synthetases; also has a role in nuclear export of tRNAs |
|
| YGR108W | 0.06 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YER136W | 0.06 |
GDI1
|
GDP dissociation inhibitor, regulates vesicle traffic in secretory pathways by regulating the dissociation of GDP from the Sec4/Ypt/rab family of GTP binding proteins |
|
| YNR054C | 0.06 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YOL086C | 0.06 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YKL043W | 0.06 |
PHD1
|
Transcriptional activator that enhances pseudohyphal growth; regulates expression of FLO11, an adhesin required for pseudohyphal filament formation; similar to StuA, an A. nidulans developmental regulator; potential Cdc28p substrate |
|
| YHL017W | 0.06 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein co-localizes with clathrin-coated vesicles |
||
| YLR420W | 0.06 |
URA4
|
Dihydroorotase, catalyzes the third enzymatic step in the de novo biosynthesis of pyrimidines, converting carbamoyl-L-aspartate into dihydroorotate |
|
| YJR010W | 0.06 |
MET3
|
ATP sulfurylase, catalyzes the primary step of intracellular sulfate activation, essential for assimilatory reduction of sulfate to sulfide, involved in methionine metabolism |
Network of associatons between targets according to the STRING database.
First level regulatory network of YPR015C
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.3 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 0.6 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.4 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.2 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.1 | 0.2 | GO:2000197 | regulation of mRNA export from nucleus(GO:0010793) regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 0.3 | GO:1903530 | regulation of exocytosis(GO:0017157) regulation of secretion(GO:0051046) regulation of secretion by cell(GO:1903530) |
| 0.1 | 0.3 | GO:0000296 | spermine transport(GO:0000296) |
| 0.1 | 0.2 | GO:0045895 | negative regulation of cellular amine metabolic process(GO:0033239) regulation of cellular amine catabolic process(GO:0033241) positive regulation of mating-type specific transcription, DNA-templated(GO:0045895) |
| 0.1 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.3 | GO:0007535 | donor selection(GO:0007535) |
| 0.1 | 0.2 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.1 | 0.1 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.1 | 0.2 | GO:0046160 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.1 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.4 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 0.4 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.7 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.2 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.0 | 0.5 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.6 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.3 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.1 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0009226 | UDP-N-acetylglucosamine metabolic process(GO:0006047) UDP-N-acetylglucosamine biosynthetic process(GO:0006048) nucleotide-sugar biosynthetic process(GO:0009226) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) heme biosynthetic process(GO:0006783) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:1900434 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.0 | 0.1 | GO:0045862 | positive regulation of proteolysis(GO:0045862) |
| 0.0 | 0.4 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.0 | 0.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.1 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:0042991 | transcription factor import into nucleus(GO:0042991) |
| 0.0 | 0.0 | GO:0090342 | regulation of cell aging(GO:0090342) regulation of replicative cell aging(GO:1900062) |
| 0.0 | 0.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.1 | GO:0070893 | DNA integration(GO:0015074) transposon integration(GO:0070893) |
| 0.0 | 0.1 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.4 | GO:0016129 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.0 | 0.2 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.2 | GO:0033673 | negative regulation of protein kinase activity(GO:0006469) negative regulation of kinase activity(GO:0033673) |
| 0.0 | 0.0 | GO:0009162 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.0 | GO:0016094 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) dolichol metabolic process(GO:0019348) dolichol biosynthetic process(GO:0019408) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.0 | GO:0032102 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.0 | 0.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.0 | 0.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0046513 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:1904666 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) regulation of ubiquitin protein ligase activity(GO:1904666) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.2 | GO:0006885 | regulation of pH(GO:0006885) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032160 | septin filament array(GO:0032160) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0043529 | GET complex(GO:0043529) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0034456 | UTP-C complex(GO:0034456) |
| 0.0 | 0.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0000399 | cellular bud neck septin structure(GO:0000399) cleavage apparatus septin structure(GO:0032161) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005724 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.0 | GO:0070209 | ASTRA complex(GO:0070209) R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0000444 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.2 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.1 | 0.3 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.1 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.5 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0001026 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0005496 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.0 | 0.1 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 0.1 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.0 | GO:0015186 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0030332 | cyclin binding(GO:0030332) ubiquitin-protein transferase activator activity(GO:0097027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.1 | 0.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.0 | REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | Genes involved in NFkB and MAP kinases activation mediated by TLR4 signaling repertoire |