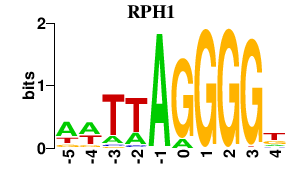
Results for RPH1
Z-value: 3.74
Transcription factors associated with RPH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RPH1
|
S000000971 | JmjC domain-containing histone demethylase |
Activity-expression correlation:
Activity profile of RPH1 motif
Sorted Z-values of RPH1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YMR107W | 57.85 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YHR096C | 54.27 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YBR072W | 53.23 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YMR175W | 48.90 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YGR087C | 47.02 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YNR034W-A | 41.20 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YKL065W-A | 40.50 |
Putative protein of unknown function |
||
| YGR256W | 34.05 |
GND2
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone |
|
| YFR017C | 33.78 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YFR017C is not an essential gene |
||
| YHR139C | 32.91 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YOL052C-A | 32.54 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YKL217W | 31.95 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YOR393W | 31.87 |
ERR1
|
Protein of unknown function, has similarity to enolases |
|
| YAL062W | 30.97 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YAL054C | 30.54 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YDR277C | 29.30 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YNL117W | 28.74 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YPL223C | 27.88 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YIL136W | 27.13 |
OM45
|
Protein of unknown function, major constituent of the mitochondrial outer membrane; located on the outer (cytosolic) face of the outer membrane |
|
| YGR088W | 26.11 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YFR053C | 24.11 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YPL036W | 23.72 |
PMA2
|
Plasma membrane H+-ATPase, isoform of Pma1p, involved in pumping protons out of the cell; regulator of cytoplasmic pH and plasma membrane potential |
|
| YPL054W | 22.97 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YKL107W | 22.47 |
Putative protein of unknown function |
||
| YPR010C-A | 21.99 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YFL011W | 21.80 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YLR312C | 21.79 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YBR241C | 21.40 |
Putative transporter, member of the sugar porter family; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar membrane; YBR241C is not an essential gene |
||
| YMR081C | 21.40 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YGR043C | 21.26 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YOR391C | 21.03 |
HSP33
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31 and S. cerevisiae Hsp31p, Hsp32p, and Sno4p; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease |
|
| YIL099W | 20.16 |
SGA1
|
Intracellular sporulation-specific glucoamylase involved in glycogen degradation; induced during starvation of a/a diploids late in sporulation, but dispensable for sporulation |
|
| YDR070C | 20.05 |
FMP16
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YBL049W | 19.89 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YMR174C | 19.80 |
PAI3
|
Cytoplasmic proteinase A (Pep4p) inhibitor, dependent on Pbs2p and Hog1p protein kinases for osmotic induction; intrinsically unstructured, N-terminal half becomes ordered in the active site of proteinase A upon contact |
|
| YMR105C | 19.53 |
PGM2
|
Phosphoglucomutase, catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose metabolism; functions as the acceptor for a Glc-phosphotransferase |
|
| YOR186W | 18.67 |
Putative protein of unknown function; proper regulation of expression during heat stress is sphingolipid-dependent |
||
| YDL085W | 18.38 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YPR030W | 18.35 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YKL103C | 18.31 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YER067W | 17.81 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YER067W is not an essential gene |
||
| YKL163W | 17.79 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YBL075C | 17.35 |
SSA3
|
ATPase involved in protein folding and the response to stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the heat shock protein 70 (HSP70) family; localized to the cytoplasm |
|
| YKL148C | 17.19 |
SDH1
|
Flavoprotein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YAL034C | 17.16 |
FUN19
|
Non-essential protein of unknown function |
|
| YEL039C | 17.01 |
CYC7
|
Cytochrome c isoform 2, expressed under hypoxic conditions; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YDL222C | 16.65 |
FMP45
|
Integral membrane protein localized to mitochondria (untagged protein) and eisosomes, immobile patches at the cortex associated with endocytosis; sporulation and sphingolipid content are altered in mutants; has homologs SUR7 and YNL194C |
|
| YHL040C | 16.58 |
ARN1
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores |
|
| YNR001C | 16.55 |
CIT1
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate; the rate-limiting enzyme of the TCA cycle; nuclear encoded mitochondrial protein |
|
| YBR117C | 16.55 |
TKL2
|
Transketolase, similar to Tkl1p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YKL102C | 16.17 |
Dubious open reading frame unlikely to encode a functional protein; deletion confers sensitivity to citric acid; predicted protein would include a thiol-disulfide oxidoreductase active site |
||
| YOR031W | 16.15 |
CRS5
|
Copper-binding metallothionein, required for wild-type copper resistance |
|
| YDL204W | 16.07 |
RTN2
|
Protein of unknown function; has similarity to mammalian reticulon proteins; member of the RTNLA (reticulon-like A) subfamily |
|
| YDL130W-A | 15.95 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YDR343C | 15.68 |
HXT6
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt7p, expressed at high basal levels relative to other HXTs, repression of expression by high glucose requires SNF3 |
|
| YIL101C | 15.58 |
XBP1
|
Transcriptional repressor that binds to promoter sequences of the cyclin genes, CYS3, and SMF2; expression is induced by stress or starvation during mitosis, and late in meiosis; member of the Swi4p/Mbp1p family; potential Cdc28p substrate |
|
| YFL058W | 15.55 |
THI5
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YJL116C | 15.41 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YAR053W | 15.33 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL210W | 15.27 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YGR067C | 15.23 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YLR327C | 14.60 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YER103W | 14.54 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YGR243W | 14.27 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YMR250W | 13.94 |
GAD1
|
Glutamate decarboxylase, converts glutamate into gamma-aminobutyric acid (GABA) during glutamate catabolism; involved in response to oxidative stress |
|
| YBR214W | 13.82 |
SDS24
|
One of two S. cerevisiae homologs (Sds23p and Sds24p) of the Schizosaccharomyces pombe Sds23 protein, which genetic studies have implicated in APC/cyclosome regulation; may play an indirect role in fluid-phase endocytosis |
|
| YNL014W | 13.80 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YHR033W | 13.64 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YLR377C | 13.58 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YML089C | 13.54 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YLR356W | 13.45 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YOR348C | 13.33 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YGR201C | 13.31 |
Putative protein of unknown function |
||
| YPL240C | 13.23 |
HSP82
|
Hsp90 chaperone required for pheromone signaling and negative regulation of Hsf1p; docks with Tom70p for mitochondrial preprotein delivery; promotes telomerase DNA binding and nucleotide addition; interacts with Cns1p, Cpr6p, Cpr7p, Sti1p |
|
| YDL218W | 12.90 |
Putative protein of unknown function; YDL218W transcription is regulated by Azf1p and induced by starvation and aerobic conditions |
||
| YOR374W | 12.85 |
ALD4
|
Mitochondrial aldehyde dehydrogenase, required for growth on ethanol and conversion of acetaldehyde to acetate; phosphorylated; activity is K+ dependent; utilizes NADP+ or NAD+ equally as coenzymes; expression is glucose repressed |
|
| YMR251W | 12.55 |
GTO3
|
Omega class glutathione transferase; putative cytosolic localization |
|
| YDR258C | 12.50 |
HSP78
|
Oligomeric mitochondrial matrix chaperone that cooperates with Ssc1p in mitochondrial thermotolerance after heat shock; prevents the aggregation of misfolded matrix proteins; component of the mitochondrial proteolysis system |
|
| YML131W | 12.07 |
Putative protein of unknown function with similarity to oxidoreductases; HOG1 and SKO1-dependent mRNA expression is induced after osmotic shock; GFP-fusion protein localizes to the cytoplasm and is induced by the DNA-damaging agent MMS |
||
| YNL194C | 11.86 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YEL070W | 11.85 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YOR346W | 11.72 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YGR144W | 11.52 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YOR392W | 11.45 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; gene expression induced by heat |
||
| YPL222W | 11.29 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YOR100C | 11.21 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YAR060C | 11.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR127W | 10.99 |
Putative protein of unknown function; expression is activated by transcription factor YRM1/YOR172W; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YOR345C | 10.87 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YMR206W | 10.87 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YIL107C | 10.85 |
PFK26
|
6-phosphofructo-2-kinase, inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, has negligible fructose-2,6-bisphosphatase activity, transcriptional regulation involves protein kinase A |
|
| YLR149C | 10.81 |
Putative protein of unknown function; YLR149C is not an essential gene |
||
| YIL086C | 10.73 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR192W | 10.67 |
AQY1
|
Spore-specific water channel that mediates the transport of water across cell membranes, developmentally controlled; may play a role in spore maturation, probably by allowing water outflow, may be involved in freeze tolerance |
|
| YKL050C | 10.43 |
Protein of unknown function; the YKL050W protein is a target of the SCFCdc4 ubiquitin ligase complex and YKL050W transcription is regulated by Azf1p |
||
| YDL214C | 10.35 |
PRR2
|
Serine/threonine protein kinase that inhibits pheromone induced signalling downstream of MAPK, possibly at the level of the Ste12p transcription factor |
|
| YPL186C | 10.25 |
UIP4
|
Protein that interacts with Ulp1p, a Ubl (ubiquitin-like protein)-specific protease for Smt3p protein conjugates; detected in a phosphorylated state in the mitochondrial outer membrane; also detected in ER and nuclear envelope |
|
| YMR181C | 10.24 |
Protein of unknown function; mRNA transcribed as part of a bicistronic transcript with a predicted transcriptional repressor RGM1/YMR182C; mRNA is destroyed by nonsense-mediated decay (NMD); YMR181C is not an essential gene |
||
| YJR154W | 10.23 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YER096W | 10.08 |
SHC1
|
Sporulation-specific activator of Chs3p (chitin synthase III), required for the synthesis of the chitosan layer of ascospores; has similarity to Skt5p, which activates Chs3p during vegetative growth; transcriptionally induced at alkaline pH |
|
| YDR342C | 10.03 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YEL009C-A | 9.98 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL156W | 9.97 |
AMS1
|
Vacuolar alpha mannosidase, involved in free oligosaccharide (fOS) degradation; delivered to the vacuole in a novel pathway separate from the secretory pathway |
|
| YOR173W | 9.94 |
DCS2
|
Non-essential, stress induced regulatory protein containing a HIT (histidine triad) motif; modulates m7G-oligoribonucleotide metabolism; inhibits Dcs1p; regulated by Msn2p, Msn4p, and the Ras-cAMP-cAPK signaling pathway, similar to Dcs1p. |
|
| YNL332W | 9.88 |
THI12
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YOL118C | 9.78 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YAL061W | 9.73 |
BDH2
|
Putative medium-chain alcohol dehydrogenase with similarity to BDH1; transcription induced by constitutively active PDR1 and PDR3; BDH2 is an essential gene |
|
| YPR184W | 9.61 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YAL005C | 9.56 |
SSA1
|
ATPase involved in protein folding and nuclear localization signal (NLS)-directed nuclear transport; member of heat shock protein 70 (HSP70) family; forms a chaperone complex with Ydj1p; localized to the nucleus, cytoplasm, and cell wall |
|
| YLR164W | 9.55 |
Mitochondrial inner membrane of unknown function; similar to Tim18p and Sdh4p; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
||
| YNL115C | 9.47 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YNL115C is not an essential gene |
||
| YJR115W | 9.45 |
Putative protein of unknown function |
||
| YGL096W | 9.44 |
TOS8
|
Homeodomain-containing transcription factor; SBF regulated target gene that in turn regulates expression of genes involved in G1/S phase events such as bud site selection, bud emergence and cell cycle progression; similarity to Cup9p |
|
| YIL055C | 9.39 |
Putative protein of unknown function |
||
| YJL089W | 9.37 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YIR039C | 9.35 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YHR139C-A | 9.34 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAL063C | 9.31 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YKL066W | 9.31 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene YNK1 |
||
| YMR196W | 9.22 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR196W is not an essential gene |
||
| YDR533C | 8.97 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YLR326W | 8.96 |
Hypothetical protein |
||
| YOL083W | 8.96 |
Hypothetical protein |
||
| YPL185W | 8.94 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene UIP4/YPL186C |
||
| YMR194C-B | 8.84 |
Putative protein of unknown function |
||
| YNL092W | 8.84 |
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family; YNL092W is not an essential gene |
||
| YMR169C | 8.81 |
ALD3
|
Cytoplasmic aldehyde dehydrogenase, involved in beta-alanine synthesis; uses NAD+ as the preferred coenzyme; very similar to Ald2p; expression is induced by stress and repressed by glucose |
|
| YBR201C-A | 8.75 |
Putative protein of unknown function |
||
| YLL041C | 8.74 |
SDH2
|
Iron-sulfur protein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YMR053C | 8.59 |
STB2
|
Protein that interacts with Sin3p in a two-hybrid assay and is part of a large protein complex with Sin3p and Stb1p |
|
| YIL100C-A | 8.59 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR267W | 8.57 |
BOP2
|
Protein of unknown function, overproduction suppresses a pam1 slv3 double null mutation |
|
| YNL180C | 8.53 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YDL181W | 8.50 |
INH1
|
Protein that inhibits ATP hydrolysis by the F1F0-ATP synthase; inhibitory function is enhanced by stabilizing proteins Stf1p and Stf2p; has similarity to Stf1p; has a calmodulin-binding motif and binds calmodulin in vitro |
|
| YML120C | 8.49 |
NDI1
|
NADH:ubiquinone oxidoreductase, transfers electrons from NADH to ubiquinone in the respiratory chain but does not pump protons, in contrast to the higher eukaryotic multisubunit respiratory complex I; phosphorylated; homolog of human AMID |
|
| YHR087W | 8.47 |
RTC3
|
Protein of unknown function involved in RNA metabolism; has structural similarity to SBDS, the human protein mutated in Shwachman-Diamond Syndrome (the yeast SBDS ortholog = SDO1); null mutation suppresses cdc13-1 temperature sensitivity |
|
| YPL271W | 8.46 |
ATP15
|
Epsilon subunit of the F1 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; phosphorylated |
|
| YML118W | 8.45 |
NGL3
|
Putative endonuclease, has a domain similar to a magnesium-dependent endonuclease motif in mRNA deadenylase Ccr4p; similar to Ngl1p and Ngl2p |
|
| YHR212C | 8.41 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR054W | 8.36 |
YRO2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in a phosphorylated state in highly purified mitochondria in high-throughput studies; transcriptionally regulated by Haa1p |
|
| YEL011W | 8.34 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YBR280C | 8.27 |
SAF1
|
F-Box protein involved in proteasome-dependent degradation of Aah1p during entry of cells into quiescence; interacts with Skp1 |
|
| YDR505C | 8.27 |
PSP1
|
Asn and gln rich protein of unknown function; high-copy suppressor of POL1 (DNA polymerase alpha) and partial suppressor of CDC2 (polymerase delta) and CDC6 (pre-RC loading factor) mutations; overexpression results in growth inhibition |
|
| YLR080W | 8.27 |
EMP46
|
Integral membrane component of endoplasmic reticulum-derived COPII-coated vesicles, which function in ER to Golgi transport |
|
| YPL119C | 8.26 |
DBP1
|
Putative ATP-dependent RNA helicase of the DEAD-box protein family; mutants show reduced stability of the 40S ribosomal subunit scanning through 5' untranslated regions of mRNAs |
|
| YML090W | 8.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YIL087C | 8.16 |
LRC2
|
Putative protein of unknown function; protein is detected in purified mitochondria in high-throughput studies; null mutant displays decreased mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YJR150C | 8.10 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YFL019C | 7.95 |
Dubious open reading frame unlikely to encode a protein; YFL019C is not an essential gene |
||
| YBR203W | 7.94 |
COS111
|
Protein required for resistance to the antifungal drug ciclopirox olamine; not related to the subtelomerically-encoded COS family; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL077W | 7.88 |
APJ1
|
Putative chaperone of the HSP40 (DNAJ) family; overexpression interferes with propagation of the [Psi+] prion; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL179C | 7.84 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YBR132C | 7.83 |
AGP2
|
High affinity polyamine permease, preferentially uses spermidine over putrescine; expression is down-regulated by osmotic stress; plasma membrane carnitine transporter, also functions as a low-affinity amino acid permease |
|
| YKL031W | 7.80 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YPR117W | 7.78 |
Putative protein of unknown function |
||
| YIL100W | 7.75 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the dubious ORF YIL100C-A |
||
| YPL230W | 7.71 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YHL021C | 7.67 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YER065C | 7.63 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YJL144W | 7.61 |
Cytoplasmic hydrophilin of unknown function, possibly involved in the dessication response; expression induced by osmotic stress, starvation and during stationary phase; GFP-fusion protein is induced by the DNA-damaging agent MMS |
||
| YPR027C | 7.61 |
Putative protein of unknown function |
||
| YKR033C | 7.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene DAL80 |
||
| YHR212W-A | 7.59 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YKL109W | 7.53 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YAL028W | 7.51 |
FRT2
|
Tail-anchored endoplasmic reticulum membrane protein, interacts with homolog Frt1p but is not a substrate of calcineurin (unlike Frt1p), promotes growth in conditions of high Na+, alkaline pH, or cell wall stress; potential Cdc28p substrate |
|
| YAL067C | 7.44 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YMR103C | 7.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR077C | 7.37 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YEL010W | 7.31 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR118C | 7.25 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YPL282C | 7.09 |
PAU22
|
Hypothetical protein |
|
| YPL017C | 7.05 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YDR043C | 7.02 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YJL020C | 7.02 |
BBC1
|
Protein possibly involved in assembly of actin patches; interacts with an actin assembly factor Las17p and with the SH3 domains of Type I myosins Myo3p and Myo5p; localized predominantly to cortical actin patches |
|
| YMR256C | 7.01 |
COX7
|
Subunit VII of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YJR149W | 6.97 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YJL103C | 6.97 |
GSM1
|
Putative zinc cluster protein of unknown function; proposed to be involved in the regulation of energy metabolism, based on patterns of expression and sequence analysis |
|
| YAR050W | 6.97 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YLR337C | 6.96 |
VRP1
|
Proline-rich actin-associated protein involved in cytoskeletal organization and cytokinesis; related to mammalian Wiskott-Aldrich syndrome protein (WASP)-interacting protein (WIP) |
|
| YKL146W | 6.95 |
AVT3
|
Vacuolar transporter, exports large neutral amino acids from the vacuole; member of a family of seven S. cerevisiae genes (AVT1-7) related to vesicular GABA-glycine transporters |
|
| YOL047C | 6.95 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YPR160W | 6.94 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YCR010C | 6.91 |
ADY2
|
Acetate transporter required for normal sporulation; phosphorylated in mitochondria |
|
| YMR170C | 6.87 |
ALD2
|
Cytoplasmic aldehyde dehydrogenase, involved in ethanol oxidation and beta-alanine biosynthesis; uses NAD+ as the preferred coenzyme; expression is stress induced and glucose repressed; very similar to Ald3p |
|
| YHR138C | 6.83 |
Putative protein of unknown function; has similarity to Pbi2p; double null mutant lacking Pbi2p and Yhr138p exhibits highly fragmented vacuoles |
||
| YCR025C | 6.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR025C is not an essential gene |
||
| YKL147C | 6.73 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene AVT3 |
||
| YJL161W | 6.73 |
FMP33
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YGR248W | 6.70 |
SOL4
|
6-phosphogluconolactonase with similarity to Sol3p |
|
| YAL039C | 6.68 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YOR065W | 6.67 |
CYT1
|
Cytochrome c1, component of the mitochondrial respiratory chain; expression is regulated by the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex |
|
| YDR322C-A | 6.64 |
TIM11
|
Subunit e of mitochondrial F1F0-ATPase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; essential for the dimeric and oligomeric state of ATP synthase |
|
| YEL030C-A | 6.62 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YML091C | 6.60 |
RPM2
|
Protein subunit of mitochondrial RNase P, has roles in nuclear transcription, cytoplasmic and mitochondrial RNA processing, and mitochondrial translation; distributed to mitochondria, cytoplasmic processing bodies, and the nucleus |
|
| YLR136C | 6.58 |
TIS11
|
mRNA-binding protein expressed during iron starvation; binds to a sequence element in the 3'-untranslated regions of specific mRNAs to mediate their degradation; involved in iron homeostasis |
|
| YDR446W | 6.56 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YMR084W | 6.55 |
Putative protein of unknown function; YMR084W and adjacent ORF YMR085W are merged in related strains |
||
| YEL008W | 6.52 |
Hypothetical protein predicted to be involved in metabolism |
||
| YPR061C | 6.52 |
JID1
|
Probable Hsp40p co-chaperone, has a DnaJ-like domain and appears to be involved in ER-associated degradation of misfolded proteins containing a tightly folded cytoplasmic domain; inhibits replication of Brome mosaic virus in S. cerevisiae |
|
| YPR002W | 6.50 |
PDH1
|
Mitochondrial protein that participates in respiration, induced by diauxic shift; homologous to E. coli PrpD, may take part in the conversion of 2-methylcitrate to 2-methylisocitrate |
Network of associatons between targets according to the STRING database.
First level regulatory network of RPH1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.6 | 50.4 | GO:0006848 | pyruvate transport(GO:0006848) |
| 12.1 | 48.6 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 12.1 | 48.3 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 10.2 | 30.5 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 9.8 | 29.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 9.2 | 27.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 8.6 | 25.8 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 8.3 | 91.1 | GO:0015758 | glucose transport(GO:0015758) |
| 8.1 | 40.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 7.3 | 36.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 7.0 | 21.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 6.6 | 19.9 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 6.4 | 32.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 6.2 | 30.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 5.9 | 17.8 | GO:0043335 | protein unfolding(GO:0043335) |
| 5.9 | 5.9 | GO:0034310 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 5.7 | 45.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 5.0 | 10.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 4.9 | 9.7 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 4.6 | 41.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 4.0 | 16.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 3.9 | 15.8 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 3.9 | 15.7 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 3.9 | 23.2 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 3.8 | 11.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 3.5 | 35.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 3.1 | 31.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 3.1 | 12.4 | GO:0015847 | putrescine transport(GO:0015847) |
| 3.1 | 15.3 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 3.0 | 8.9 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 2.9 | 26.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 2.8 | 8.5 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 2.8 | 44.7 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 2.6 | 15.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 2.6 | 10.3 | GO:0043901 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 2.5 | 40.7 | GO:0042724 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 2.5 | 12.4 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 2.5 | 12.4 | GO:0032974 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 2.3 | 7.0 | GO:0071467 | cellular response to pH(GO:0071467) |
| 2.3 | 14.0 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 2.2 | 6.7 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 2.2 | 13.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 2.2 | 17.5 | GO:0015891 | siderophore transport(GO:0015891) |
| 2.2 | 6.5 | GO:0019626 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 2.1 | 8.4 | GO:0032102 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 2.0 | 26.6 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 2.0 | 7.9 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 1.9 | 17.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 1.9 | 29.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 1.9 | 5.6 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 1.8 | 5.3 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 1.7 | 7.0 | GO:0019320 | hexose catabolic process(GO:0019320) |
| 1.7 | 8.7 | GO:0019405 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 1.7 | 13.5 | GO:0072593 | reactive oxygen species metabolic process(GO:0072593) |
| 1.7 | 3.3 | GO:0072353 | age-dependent response to reactive oxygen species(GO:0001315) cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 1.6 | 6.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 1.5 | 1.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 1.5 | 3.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 1.5 | 1.5 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 1.5 | 3.0 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 1.5 | 2.9 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 1.5 | 7.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 1.5 | 2.9 | GO:0001080 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 1.4 | 15.9 | GO:0015696 | ammonium transport(GO:0015696) |
| 1.4 | 4.3 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.4 | 12.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 1.4 | 5.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 1.4 | 5.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 1.3 | 4.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.3 | 7.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 1.3 | 5.3 | GO:0000431 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 1.3 | 2.5 | GO:0000411 | regulation of transcription by galactose(GO:0000409) positive regulation of transcription by galactose(GO:0000411) |
| 1.2 | 8.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 1.2 | 7.4 | GO:0000255 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) |
| 1.2 | 27.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 1.2 | 10.5 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 1.1 | 3.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 1.1 | 1.1 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 1.1 | 2.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 1.1 | 21.7 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 1.1 | 2.2 | GO:0071456 | response to hypoxia(GO:0001666) cellular response to hypoxia(GO:0071456) |
| 1.1 | 4.3 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 1.1 | 4.2 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 1.0 | 2.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 1.0 | 4.0 | GO:0051303 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 1.0 | 17.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 1.0 | 7.9 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 1.0 | 4.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 1.0 | 2.9 | GO:0015888 | thiamine transport(GO:0015888) |
| 1.0 | 40.2 | GO:0071940 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 1.0 | 2.9 | GO:1903137 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.9 | 2.8 | GO:0044109 | cellular alcohol catabolic process(GO:0044109) |
| 0.9 | 3.7 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.9 | 4.6 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.9 | 2.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.9 | 1.8 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.9 | 0.9 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.9 | 6.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.9 | 7.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.8 | 0.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.8 | 5.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.8 | 4.9 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.8 | 2.4 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.8 | 3.9 | GO:0001881 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.8 | 4.7 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.8 | 3.1 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.8 | 3.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.8 | 9.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.7 | 2.2 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.7 | 4.3 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.7 | 0.7 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.7 | 4.8 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.7 | 2.7 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.7 | 3.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.7 | 2.0 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.6 | 9.5 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.6 | 2.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.6 | 7.7 | GO:0007135 | meiosis II(GO:0007135) meiotic sister chromatid segregation(GO:0045144) |
| 0.6 | 2.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.6 | 1.7 | GO:2000911 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.6 | 6.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.6 | 3.4 | GO:0007532 | regulation of mating-type specific transcription, DNA-templated(GO:0007532) |
| 0.6 | 2.2 | GO:0005977 | glycogen metabolic process(GO:0005977) energy reserve metabolic process(GO:0006112) |
| 0.6 | 23.1 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.5 | 0.5 | GO:0045723 | response to cold(GO:0009409) positive regulation of fatty acid biosynthetic process(GO:0045723) cellular response to cold(GO:0070417) |
| 0.5 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.5 | 1.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 2.0 | GO:0034033 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.5 | 1.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.5 | 2.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.5 | 3.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.5 | 4.7 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.5 | 2.3 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.5 | 1.8 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.4 | 1.8 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.4 | 8.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.4 | 0.9 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.4 | 4.6 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.4 | 1.3 | GO:0032079 | positive regulation of nuclease activity(GO:0032075) positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.4 | 1.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.4 | 3.1 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.4 | 1.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.4 | 15.6 | GO:0048468 | ascospore formation(GO:0030437) cell development(GO:0048468) |
| 0.4 | 3.6 | GO:0000767 | cell morphogenesis involved in conjugation(GO:0000767) |
| 0.4 | 1.4 | GO:0046503 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) glycerolipid catabolic process(GO:0046503) |
| 0.3 | 1.7 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.3 | 6.5 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.3 | 1.7 | GO:0051598 | meiotic cell cycle checkpoint(GO:0033313) meiotic recombination checkpoint(GO:0051598) |
| 0.3 | 1.0 | GO:0071050 | snoRNA polyadenylation(GO:0071050) |
| 0.3 | 0.3 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.3 | 1.0 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.3 | 1.6 | GO:0046033 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.3 | 1.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 2.9 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.3 | 0.3 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.3 | 1.8 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.3 | 0.6 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.3 | 2.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 0.6 | GO:0015883 | FAD transport(GO:0015883) |
| 0.3 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 4.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.3 | 1.5 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.3 | 6.9 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.2 | 2.9 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 0.5 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.2 | 2.4 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.2 | 0.7 | GO:0034762 | regulation of transmembrane transport(GO:0034762) regulation of ion transport(GO:0043269) |
| 0.2 | 2.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.2 | 1.1 | GO:0000078 | obsolete cytokinesis after mitosis checkpoint(GO:0000078) |
| 0.2 | 1.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.2 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 2.5 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.2 | 0.7 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.2 | 2.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 0.7 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) |
| 0.2 | 1.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.2 | 0.2 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.2 | 0.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.2 | 7.1 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.2 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.3 | GO:0051984 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of mitotic nuclear division(GO:0045840) positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of proteolysis(GO:0045862) positive regulation of chromosome segregation(GO:0051984) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.2 | 2.4 | GO:0000902 | cell morphogenesis(GO:0000902) |
| 0.2 | 11.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.2 | 2.0 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.1 | 0.3 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 1.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.7 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.3 | GO:0035690 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.1 | 0.5 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.8 | GO:0000080 | mitotic G1 phase(GO:0000080) G1 phase(GO:0051318) |
| 0.1 | 1.0 | GO:0070272 | proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.5 | GO:0048279 | vesicle fusion with endoplasmic reticulum(GO:0048279) |
| 0.1 | 0.4 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.1 | 0.3 | GO:0018209 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.1 | 2.2 | GO:1902534 | membrane invagination(GO:0010324) lysosomal microautophagy(GO:0016237) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.2 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.5 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 0.5 | GO:0016114 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.7 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.3 | GO:0031930 | mitochondria-nucleus signaling pathway(GO:0031930) |
| 0.1 | 0.3 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 0.5 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.1 | 1.1 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 0.2 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.1 | 0.3 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.1 | 1.1 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.2 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.6 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.2 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.2 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.9 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0061393 | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061393) |
| 0.0 | 1.1 | GO:0048856 | anatomical structure morphogenesis(GO:0009653) anatomical structure development(GO:0048856) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.1 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 6.2 | 30.9 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 3.7 | 7.3 | GO:0032126 | eisosome(GO:0032126) |
| 3.3 | 9.9 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 2.9 | 35.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 2.8 | 36.8 | GO:0005619 | ascospore wall(GO:0005619) |
| 2.4 | 21.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 2.2 | 27.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 2.2 | 15.7 | GO:0034657 | GID complex(GO:0034657) |
| 2.1 | 8.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 1.9 | 33.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 1.8 | 14.3 | GO:0042597 | periplasmic space(GO:0042597) |
| 1.8 | 5.3 | GO:0001400 | mating projection base(GO:0001400) |
| 1.7 | 21.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 1.6 | 9.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 1.5 | 12.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.5 | 4.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 1.5 | 14.6 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 1.4 | 2.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 1.2 | 5.9 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 1.1 | 23.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 1.1 | 5.3 | GO:0034448 | EGO complex(GO:0034448) |
| 1.0 | 3.1 | GO:0045252 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial oxoglutarate dehydrogenase complex(GO:0009353) dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 1.0 | 4.0 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.9 | 3.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 3.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.8 | 4.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.7 | 7.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.7 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.6 | 2.6 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.6 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.6 | 1.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.6 | 8.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.6 | 12.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.6 | 17.4 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.6 | 9.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.5 | 1.6 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.5 | 23.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.5 | 2.5 | GO:0070772 | PAS complex(GO:0070772) |
| 0.5 | 9.7 | GO:0098562 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.5 | 1.8 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.5 | 64.2 | GO:0000323 | storage vacuole(GO:0000322) lytic vacuole(GO:0000323) fungal-type vacuole(GO:0000324) |
| 0.4 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 0.9 | GO:0044284 | mitochondrial crista junction(GO:0044284) |
| 0.4 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 1.7 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.4 | 323.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.4 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.4 | 6.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.4 | 7.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 5.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.3 | 1.9 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.3 | 0.9 | GO:0070274 | RES complex(GO:0070274) |
| 0.3 | 1.1 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.3 | 3.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.3 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.8 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.2 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 3.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 5.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.6 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.1 | 1.0 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.1 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.5 | GO:0048188 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0042763 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.0 | GO:0005822 | inner plaque of spindle pole body(GO:0005822) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 48.3 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 10.9 | 32.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 10.8 | 32.3 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 10.2 | 30.5 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 9.9 | 49.4 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 9.0 | 27.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 6.5 | 25.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 6.3 | 25.2 | GO:0004396 | hexokinase activity(GO:0004396) |
| 6.2 | 30.9 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 6.2 | 18.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 6.0 | 17.9 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 5.8 | 23.2 | GO:0005537 | mannose binding(GO:0005537) |
| 5.3 | 32.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 5.2 | 20.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 5.1 | 25.7 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 4.7 | 65.4 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 4.4 | 26.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 4.2 | 16.9 | GO:0015343 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 4.0 | 31.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 3.9 | 15.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 3.0 | 51.7 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 2.9 | 11.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 2.8 | 8.3 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 2.4 | 4.8 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 2.4 | 26.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 2.3 | 6.9 | GO:0015188 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 2.3 | 16.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 2.1 | 6.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 2.1 | 6.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 2.1 | 10.3 | GO:0017022 | myosin binding(GO:0017022) |
| 2.0 | 10.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 1.9 | 9.7 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 1.9 | 11.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 1.9 | 3.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 1.9 | 15.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.9 | 5.6 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 1.8 | 22.1 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 1.8 | 19.8 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 1.7 | 6.7 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 1.7 | 8.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 1.6 | 4.8 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 1.6 | 30.5 | GO:0050661 | NADP binding(GO:0050661) |
| 1.6 | 121.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 1.4 | 4.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.4 | 7.0 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 1.4 | 17.7 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 1.3 | 3.9 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 1.3 | 14.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.3 | 21.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 1.3 | 10.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 1.2 | 3.7 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 1.2 | 9.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 1.2 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 1.1 | 4.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.1 | 6.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 1.1 | 6.5 | GO:0051213 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) dioxygenase activity(GO:0051213) |
| 1.1 | 2.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 1.1 | 3.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.0 | 26.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 1.0 | 15.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 1.0 | 6.1 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 1.0 | 3.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 1.0 | 6.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.9 | 6.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.9 | 13.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.8 | 2.5 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.8 | 21.0 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.8 | 2.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.8 | 3.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.8 | 9.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.8 | 11.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.8 | 4.7 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.8 | 3.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.8 | 2.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.7 | 3.7 | GO:0016830 | carbon-carbon lyase activity(GO:0016830) |
| 0.7 | 3.4 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.7 | 1.3 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.7 | 2.0 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.6 | 1.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 3.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 1.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.6 | 5.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.5 | 1.6 | GO:0015294 | solute:cation symporter activity(GO:0015294) |
| 0.5 | 3.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.5 | 3.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.5 | 7.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.5 | 8.4 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.5 | 4.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 1.4 | GO:0008897 | holo-[acyl-carrier-protein] synthase activity(GO:0008897) |
| 0.5 | 2.7 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.4 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.4 | 3.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.4 | 1.3 | GO:0097027 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.4 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.4 | 1.6 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.4 | 3.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.4 | 8.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.4 | 4.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.4 | 1.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 2.9 | GO:0017136 | NAD-dependent histone deacetylase activity(GO:0017136) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.4 | 2.9 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.4 | 1.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 0.7 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) |
| 0.3 | 1.0 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.3 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.3 | 12.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.3 | 1.5 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.3 | 2.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 4.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 0.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.3 | 1.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.3 | 1.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 3.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 2.3 | GO:0016723 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions(GO:0016722) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.3 | 9.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 3.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.2 | 2.4 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.2 | 19.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.2 | 3.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 2.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 1.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 1.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 15.2 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.2 | 1.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 0.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.2 | 0.9 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.2 | 2.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.2 | 0.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.2 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.7 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 4.6 | GO:0043492 | ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.3 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 1.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.1 | GO:0099600 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.1 | 1.4 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 1.7 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 1.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.3 | GO:0000772 | mating pheromone activity(GO:0000772) receptor binding(GO:0005102) pheromone activity(GO:0005186) |
| 0.1 | 0.9 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 4.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 0.2 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.0 | 0.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 3.1 | 15.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 1.8 | 7.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 1.1 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.7 | 2.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.7 | 2.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.7 | 2.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.3 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 453.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.3 | 1.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 3.0 | 33.5 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 1.3 | 2.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.9 | 2.7 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.9 | 3.4 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.5 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.5 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.4 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 5.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 453.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 0.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 1.4 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |