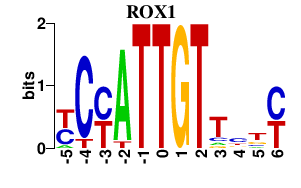
Results for ROX1
Z-value: 1.18
Transcription factors associated with ROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ROX1
|
S000006269 | Heme-dependent repressor of hypoxic genes |
Activity-expression correlation:
Activity profile of ROX1 motif
Sorted Z-values of ROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFR055W | 18.16 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YFR056C | 16.76 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YOL086C | 10.73 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YCR018C | 7.54 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YDR033W | 7.50 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YKR038C | 7.37 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YNR001W-A | 7.32 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YHR181W | 6.73 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YDR044W | 6.67 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YGR108W | 6.34 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YNL111C | 5.44 |
CYB5
|
Cytochrome b5, involved in the sterol and lipid biosynthesis pathways; acts as an electron donor to support sterol C5-6 desaturation |
|
| YOL056W | 5.00 |
GPM3
|
Homolog of Gpm1p phosphoglycerate mutase, which converts 3-phosphoglycerate to 2-phosphoglycerate in glycolysis; may be non-functional derivative of a gene duplication event |
|
| YKL218C | 4.99 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YHR180W-A | 4.95 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YER011W | 4.65 |
TIR1
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expression is downregulated at acidic pH and induced by cold shock and anaerobiosis; abundance is increased in cells cultured without shaking |
|
| YOL085C | 4.48 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YFR054C | 4.45 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR226C | 4.42 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YHR007C | 4.31 |
ERG11
|
Lanosterol 14-alpha-demethylase, catalyzes the C-14 demethylation of lanosterol to form 4,4''-dimethyl cholesta-8,14,24-triene-3-beta-ol in the ergosterol biosynthesis pathway; member of the cytochrome P450 family |
|
| YDR385W | 4.19 |
EFT2
|
Elongation factor 2 (EF-2), also encoded by EFT1; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YPR063C | 4.17 |
ER-localized protein of unknown function |
||
| YMR199W | 3.97 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YBR238C | 3.96 |
Mitochondrial membrane protein with similarity to Rmd9p; not required for respiratory growth but causes a synthetic respiratory defect in combination with rmd9 mutations; transcriptionally up-regulated by TOR; deletion increases life span |
||
| YDR509W | 3.80 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR315W | 3.74 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YMR119W | 3.69 |
ASI1
|
Putative integral membrane E3 ubiquitin ligase; acts with Asi2p and Asi3p to ensure the fidelity of SPS-sensor signalling by maintaining the dormant repressed state of gene expression in the absence of inducing signals |
|
| YGR214W | 3.66 |
RPS0A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Bp; required for maturation of 18S rRNA along with Rps0Bp; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YLR413W | 3.66 |
Putative protein of unknown function; YLR413W is not an essential gene |
||
| YLR355C | 3.59 |
ILV5
|
Acetohydroxyacid reductoisomerase, mitochondrial protein involved in branched-chain amino acid biosynthesis, also required for maintenance of wild-type mitochondrial DNA and found in mitochondrial nucleoids |
|
| YIL118W | 3.59 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YDR508C | 3.58 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YMR246W | 3.51 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YNR054C | 3.51 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YPL250W-A | 3.46 |
Identified by fungal homology and RT-PCR |
||
| YOR342C | 3.45 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YJL157C | 3.43 |
FAR1
|
Cyclin-dependent kinase inhibitor that mediates cell cycle arrest in response to pheromone; also forms a complex with Cdc24p, Ste4p, and Ste18p that may specify the direction of polarized growth during mating; potential Cdc28p substrate |
|
| YLR154C | 3.42 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YGL179C | 3.34 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YDR002W | 3.34 |
YRB1
|
Ran GTPase binding protein; involved in nuclear protein import and RNA export, ubiquitin-mediated protein degradation during the cell cycle; shuttles between the nucleus and cytoplasm; is essential; homolog of human RanBP1 |
|
| YIR021W | 3.31 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YKR092C | 3.30 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YKL182W | 3.26 |
FAS1
|
Beta subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids; contains acetyltransacylase, dehydratase, enoyl reductase, malonyl transacylase, and palmitoyl transacylase activities |
|
| YML111W | 3.12 |
BUL2
|
Component of the Rsp5p E3-ubiquitin ligase complex, involved in intracellular amino acid permease sorting, functions in heat shock element mediated gene expression, essential for growth in stress conditions, functional homolog of BUL1 |
|
| YLL045C | 3.12 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YMR032W | 3.09 |
HOF1
|
Bud neck-localized, SH3 domain-containing protein required for cytokinesis; regulates actomyosin ring dynamics and septin localization; interacts with the formins, Bni1p and Bnr1p, and with Cyk3p, Vrp1p, and Bni5p |
|
| YDR384C | 3.08 |
ATO3
|
Plasma membrane protein, regulation pattern suggests a possible role in export of ammonia from the cell; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family of putative transporters |
|
| YAL029C | 3.08 |
MYO4
|
One of two type V myosin motors (along with MYO2) involved in actin-based transport of cargos; required for mRNA transport, including ASH1 mRNA, and facilitating the growth and movement of ER tubules into the growing bud along with She3p |
|
| YNR018W | 3.02 |
AIM38
|
Putative protein of unknown function; non-tagged protein is detected in purified mitochondria; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YJL011C | 2.97 |
RPC17
|
RNA polymerase III subunit C17; physically interacts with C31, C11, and TFIIIB70; may be involved in the recruitment of pol III by the preinitiation complex |
|
| YOR369C | 2.96 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YDR344C | 2.95 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR029W | 2.93 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YGR040W | 2.85 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YKL096W-A | 2.85 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YJR047C | 2.82 |
ANB1
|
Translation initiation factor eIF-5A, promotes formation of the first peptide bond; similar to and functionally redundant with Hyp2p; undergoes an essential hypusination modification; expressed under anaerobic conditions |
|
| YMR106C | 2.81 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YBR087W | 2.81 |
RFC5
|
Subunit of heteropentameric Replication factor C (RF-C), which is a DNA binding protein and ATPase that acts as a clamp loader of the proliferating cell nuclear antigen (PCNA) processivity factor for DNA polymerases delta and epsilon |
|
| YLR448W | 2.81 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YMR177W | 2.79 |
MMT1
|
Putative metal transporter involved in mitochondrial iron accumulation; closely related to Mmt2p |
|
| YDR279W | 2.77 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YML075C | 2.75 |
HMG1
|
One of two isozymes of HMG-CoA reductase that catalyzes the conversion of HMG-CoA to mevalonate, which is a rate-limiting step in sterol biosynthesis; localizes to the nuclear envelope; overproduction induces the formation of karmellae |
|
| YOR028C | 2.73 |
CIN5
|
Basic leucine zipper transcriptional factor of the yAP-1 family that mediates pleiotropic drug resistance and salt tolerance; localizes constitutively to the nucleus |
|
| YBL092W | 2.72 |
RPL32
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L32 ribosomal protein; overexpression disrupts telomeric silencing |
|
| YDR497C | 2.71 |
ITR1
|
Myo-inositol transporter with strong similarity to the minor myo-inositol transporter Itr2p, member of the sugar transporter superfamily; expression is repressed by inositol and choline via Opi1p and derepressed via Ino2p and Ino4p |
|
| YER131W | 2.68 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YML024W | 2.65 |
RPS17A
|
Ribosomal protein 51 (rp51) of the small (40s) subunit; nearly identical to Rps17Bp and has similarity to rat S17 ribosomal protein |
|
| YHR052W | 2.64 |
CIC1
|
Essential protein that interacts with proteasome components and has a potential role in proteasome substrate specificity; also copurifies with 66S pre-ribosomal particles |
|
| YKL152C | 2.61 |
GPM1
|
Tetrameric phosphoglycerate mutase, mediates the conversion of 3-phosphoglycerate to 2-phosphoglycerate during glycolysis and the reverse reaction during gluconeogenesis |
|
| YNR013C | 2.57 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YBR011C | 2.57 |
IPP1
|
Cytoplasmic inorganic pyrophosphatase (PPase), catalyzes the rapid exchange of oxygens from Pi with water, highly expressed and essential for viability, active-site residues show identity to those from E. coli PPase |
|
| YGL039W | 2.54 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YIL053W | 2.49 |
RHR2
|
Constitutively expressed isoform of DL-glycerol-3-phosphatase; involved in glycerol biosynthesis, induced in response to both anaerobic and, along with the Hor2p/Gpp2p isoform, osmotic stress |
|
| YNL069C | 2.47 |
RPL16B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Ap, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YBR202W | 2.45 |
MCM7
|
Component of the hexameric MCM complex, which is important for priming origins of DNA replication in G1 and becomes an active ATP-dependent helicase that promotes DNA melting and elongation when activated by Cdc7p-Dbf4p in S-phase |
|
| YMR049C | 2.41 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YER137C | 2.41 |
Putative protein of unknown function |
||
| YDR345C | 2.37 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YIL011W | 2.37 |
TIR3
|
Cell wall mannoprotein of the Srp1p/Tip1p family of serine-alanine-rich proteins; expressed under anaerobic conditions and required for anaerobic growth |
|
| YOR332W | 2.35 |
VMA4
|
Subunit E of the eight-subunit V1 peripheral membrane domain of the vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane |
|
| YJL188C | 2.35 |
BUD19
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 88% of ORF overlaps the verified gene RPL39; diploid mutant displays a weak budding pattern phenotype in a systematic assay |
|
| YDR417C | 2.35 |
Hypothetical protein |
||
| YEL054C | 2.35 |
RPL12A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Bp; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YLR154W-B | 2.35 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YOR008C-A | 2.31 |
Putative protein of unknown function, includes a potential transmembrane domain; deletion results in slightly lengthened telomeres |
||
| YIL111W | 2.30 |
COX5B
|
Subunit Vb of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during anaerobic growth while its isoform Va (Cox5Ap) is expressed during aerobic growth |
|
| YLR154W-A | 2.28 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YDR037W | 2.24 |
KRS1
|
Lysyl-tRNA synthetase; also identified as a negative regulator of general control of amino acid biosynthesis |
|
| YBR265W | 2.23 |
TSC10
|
3-ketosphinganine reductase, catalyzes the second step in phytosphingosine synthesis, essential for growth in the absence of exogenous dihydrosphingosine or phytosphingosine, member of short chain dehydrogenase/reductase protein family |
|
| YOL014W | 2.22 |
Putative protein of unknown function |
||
| YGR138C | 2.21 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YBR084C-A | 2.21 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YJR123W | 2.19 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YHR208W | 2.18 |
BAT1
|
Mitochondrial branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during logarithmic phase and repressed during stationary phase |
|
| YJL153C | 2.17 |
INO1
|
Inositol 1-phosphate synthase, involved in synthesis of inositol phosphates and inositol-containing phospholipids; transcription is coregulated with other phospholipid biosynthetic genes by Ino2p and Ino4p, which bind the UASINO DNA element |
|
| YDR112W | 2.16 |
IRC2
|
Dubious open reading frame, unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR111C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YHR182C-A | 2.16 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR015C | 2.13 |
ERG5
|
C-22 sterol desaturase, a cytochrome P450 enzyme that catalyzes the formation of the C-22(23) double bond in the sterol side chain in ergosterol biosynthesis; may be a target of azole antifungal drugs |
|
| YPR170W-B | 2.13 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YGR176W | 2.13 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL014W | 2.12 |
NOP1
|
Nucleolar protein, component of the small subunit processome complex, which is required for processing of pre-18S rRNA; has similarity to mammalian fibrillarin |
|
| YAL038W | 2.11 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YGR140W | 2.11 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YHR183W | 2.11 |
GND1
|
6-phosphogluconate dehydrogenase (decarboxylating), catalyzes an NADPH regenerating reaction in the pentose phosphate pathway; required for growth on D-glucono-delta-lactone and adaptation to oxidative stress |
|
| YMR083W | 2.10 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YBR210W | 2.10 |
ERV15
|
Protein involved in export of proteins from the endoplasmic reticulum, has similarity to Erv14p |
|
| YMR208W | 2.10 |
ERG12
|
Mevalonate kinase, acts in the biosynthesis of isoprenoids and sterols, including ergosterol, from mevalonate |
|
| YBR104W | 2.07 |
YMC2
|
Mitochondrial protein, putative inner membrane transporter with a role in oleate metabolism and glutamate biosynthesis; member of the mitochondrial carrier (MCF) family; has similarity with Ymc1p |
|
| YBR249C | 2.07 |
ARO4
|
3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, catalyzes the first step in aromatic amino acid biosynthesis and is feedback-inhibited by tyrosine or high concentrations of phenylalanine or tryptophan |
|
| YDR465C | 2.04 |
RMT2
|
Arginine methyltransferase; ribosomal protein L12 is a substrate |
|
| YCR006C | 2.03 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL200C | 2.03 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YPL243W | 2.02 |
SRP68
|
Core component of the signal recognition particle (SRP) ribonucleoprotein (RNP) complex that functions in targeting nascent secretory proteins to the endoplasmic reticulum (ER) membrane |
|
| YCL018W | 2.01 |
LEU2
|
Beta-isopropylmalate dehydrogenase (IMDH), catalyzes the third step in the leucine biosynthesis pathway |
|
| YLR257W | 1.99 |
Putative protein of unknown function |
||
| YPR170W-A | 1.99 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; identified by expression profiling and mass spectrometry |
||
| YNL289W | 1.97 |
PCL1
|
Pho85 cyclin of the Pcl1,2-like subfamily, involved in entry into the mitotic cell cycle and regulation of morphogenesis, localizes to sites of polarized cell growth |
|
| YGL158W | 1.96 |
RCK1
|
Protein kinase involved in the response to oxidative stress; identified as suppressor of S. pombe cell cycle checkpoint mutations |
|
| YJR070C | 1.95 |
LIA1
|
Deoxyhypusine hydroxylase, a HEAT-repeat containing metalloenzyme that catalyses hypusine formation; binds to and is required for the modification of Hyp2p (eIF5A); complements S. pombe mmd1 mutants defective in mitochondrial positioning |
|
| YJR071W | 1.92 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YCR016W | 1.92 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus and nucleus; YCR016W is not an essential gene |
||
| YJR057W | 1.90 |
CDC8
|
Thymidylate and uridylate kinase, functions in de novo biosynthesis of pyrimidine deoxyribonucleotides; converts dTMP to dTDP and dUMP to dUTP; essential for mitotic and meiotic DNA replication; homologous to S. pombe Tmp1p |
|
| YLR198C | 1.90 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene SIK1/YLR197W |
||
| YOR153W | 1.90 |
PDR5
|
Plasma membrane ATP-binding cassette (ABC) transporter, short-lived multidrug transporter actively regulated by Pdr1p; also involved in steroid transport, cation resistance, and cellular detoxification during exponential growth |
|
| YMR194C-A | 1.88 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR126W-A | 1.87 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YDL205C | 1.87 |
HEM3
|
Phorphobilinogen deaminase, catalyzes the conversion of 4-porphobilinogen to hydroxymethylbilane, the third step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus; expression is regulated by Hap2p-Hap3p |
|
| YOR096W | 1.85 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YGL225W | 1.84 |
VRG4
|
Golgi GDP-mannose transporter; regulates Golgi function and glycosylation in Golgi |
|
| YJL090C | 1.84 |
DPB11
|
Subunit of DNA Polymerase II Epsilon complex; has BRCT domain, required on the prereplicative complex at replication origins for loading DNA polymerases to initiate DNA synthesis, also required for S/M checkpoint control |
|
| YOL149W | 1.83 |
DCP1
|
Subunit of the Dcp1p-Dcp2p decapping enzyme complex, which removes the 5' cap structure from mRNAs prior to their degradation; enhances the activity of catalytic subunit Dcp2p; regulated by DEAD box protein Dhh1p |
|
| YPR102C | 1.82 |
RPL11A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl11Bp; involved in ribosomal assembly; depletion causes degradation of proteins and RNA of the 60S subunit; has similarity to E. coli L5 and rat L11 |
|
| YOL124C | 1.81 |
TRM11
|
Catalytic subunit of an adoMet-dependent tRNA methyltransferase complex (Trm11p-Trm112p), required for the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs; contains a THUMP domain and a methyltransferase domain |
|
| YLR180W | 1.81 |
SAM1
|
S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) |
|
| YJL008C | 1.80 |
CCT8
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, required for the assembly of actin and tubulins in vivo |
|
| YEL051W | 1.78 |
VMA8
|
Subunit D of the eight-subunit V1 peripheral membrane domain of the vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; plays a role in the coupling of proton transport and ATP hydrolysis |
|
| YNR003C | 1.76 |
RPC34
|
RNA polymerase III subunit C34; interacts with TFIIIB70 and is a key determinant in pol III recruitment by the preinitiation complex |
|
| YMR205C | 1.75 |
PFK2
|
Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes |
|
| YPL256C | 1.75 |
CLN2
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YKL219W | 1.74 |
COS9
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YGL021W | 1.74 |
ALK1
|
Protein kinase; accumulation and phosphorylation are periodic during the cell cycle; phosphorylated in response to DNA damage; contains characteristic motifs for degradation via the APC pathway; similar to Alk2p and to mammalian haspins |
|
| YER012W | 1.73 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YGR086C | 1.72 |
PIL1
|
Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways; detected in phosphorylated state in mitochondria |
|
| YLR312W-A | 1.70 |
MRPL15
|
Mitochondrial ribosomal protein of the large subunit |
|
| YER109C | 1.69 |
FLO8
|
Transcription factor required for flocculation, diploid filamentous growth, and haploid invasive growth; genome reference strain S288C and most laboratory strains have a mutation in this gene |
|
| YDR091C | 1.68 |
RLI1
|
Essential iron-sulfur protein required for ribosome biogenesis and translation initiation; facilitates binding of a multifactor complex (MFC) of translation initiation factors to the small ribosomal subunit; predicted ABC family ATPase |
|
| YGL001C | 1.66 |
ERG26
|
C-3 sterol dehydrogenase, catalyzes the second of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis |
|
| YKL212W | 1.66 |
SAC1
|
Phosphatidylinositol (PI) phosphatase, involved in hydrolysis of PI 3-phosphate, PI 4-phosphate and PI 3,5-bisphosphate to PI; membrane protein localizes to ER and Golgi; involved in protein trafficking, secretion and inositol metabolism |
|
| YCR019W | 1.66 |
MAK32
|
Protein necessary for structural stability of L-A double-stranded RNA-containing particles |
|
| YER102W | 1.65 |
RPS8B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps8Ap and has similarity to rat S8 ribosomal protein |
|
| YKL081W | 1.65 |
TEF4
|
Translation elongation factor EF-1 gamma |
|
| YGL157W | 1.65 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YIL074C | 1.64 |
SER33
|
3-phosphoglycerate dehydrogenase, catalyzes the first step in serine and glycine biosynthesis; isozyme of Ser3p |
|
| YLL044W | 1.63 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; transcription of both YLL044W and the overlapping gene RPL8B is reduced in the gcr1 null mutant |
||
| YOR254C | 1.63 |
SEC63
|
Essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER |
|
| YEL053W-A | 1.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YEL054C |
||
| YNL178W | 1.62 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YDL047W | 1.61 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YDR086C | 1.61 |
SSS1
|
Subunit of the Sec61p translocation complex (Sec61p-Sss1p-Sbh1p) that forms a channel for passage of secretory proteins through the endoplasmic reticulum membrane, and of the Ssh1p complex (Ssh1p-Sbh2p-Sss1p); interacts with Ost4p and Wbp1p |
|
| YEL027W | 1.61 |
CUP5
|
Proteolipid subunit of the vacuolar H(+)-ATPase V0 sector (subunit c; dicyclohexylcarbodiimide binding subunit); required for vacuolar acidification and important for copper and iron metal ion homeostasis |
|
| YIL148W | 1.61 |
RPL40A
|
Fusion protein, identical to Rpl40Bp, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YKR043C | 1.61 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YKL030W | 1.60 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene MAE1 |
||
| YOR030W | 1.60 |
DFG16
|
Probable multiple transmembrane protein, involved in diploid invasive and pseudohyphal growth upon nitrogen starvation; required for accumulation of processed Rim101p |
|
| YOR011W | 1.60 |
AUS1
|
Transporter of the ATP-binding cassette family, involved in uptake of sterols and anaerobic growth |
|
| YGL148W | 1.59 |
ARO2
|
Bifunctional chorismate synthase and flavin reductase, catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate (EPSP) to form chorismate, which is a precursor to aromatic amino acids |
|
| YEL029C | 1.59 |
BUD16
|
Putative pyridoxal kinase, key enzyme in vitamin B6 metabolism; involved in maintaining levels of pyridoxal 5'-phosphate, the active form of vitamin B6; required for genome integrity; homolog of E. coli PdxK; involved in bud-site selection |
|
| YLR132C | 1.58 |
Protein of unknown function; green fluorescent protein (GFP)-tagged protein localizes to mitochondria and nucleus; YLR132C is an essential gene |
||
| YLR110C | 1.58 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YKR026C | 1.58 |
GCN3
|
Alpha subunit of the translation initiation factor eIF2B, the guanine-nucleotide exchange factor for eIF2; activity subsequently regulated by phosphorylated eIF2; first identified as a positive regulator of GCN4 expression |
|
| YGR251W | 1.58 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YER070W | 1.57 |
RNR1
|
One of two large regulatory subunits of ribonucleotide-diphosphate reductase; the RNR complex catalyzes rate-limiting step in dNTP synthesis, regulated by DNA replication and DNA damage checkpoint pathways via localization of small subunits |
|
| YDR146C | 1.57 |
SWI5
|
Transcription factor that activates transcription of genes expressed at the M/G1 phase boundary and in G1 phase; localization to the nucleus occurs during G1 and appears to be regulated by phosphorylation by Cdc28p kinase |
|
| YGL040C | 1.57 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YLL027W | 1.57 |
ISA1
|
Mitochondrial matrix protein involved in biogenesis of the iron-sulfur (Fe/S) cluster of Fe/S proteins, isa1 deletion causes loss of mitochondrial DNA and respiratory deficiency; depletion reduces growth on nonfermentable carbon sources |
|
| YHR062C | 1.56 |
RPP1
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YMR321C | 1.56 |
Putative protein of unknown function |
||
| YOR310C | 1.56 |
NOP58
|
Protein involved in pre-rRNA processing, 18S rRNA synthesis, and snoRNA synthesis; component of the small subunit processome complex, which is required for processing of pre-18S rRNA |
|
| YHR203C | 1.54 |
RPS4B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps4Ap and has similarity to rat S4 ribosomal protein |
|
| YOR206W | 1.54 |
NOC2
|
Protein that forms a nucleolar complex with Mak21p that binds to 90S and 66S pre-ribosomes, as well as a nuclear complex with Noc3p that binds to 66S pre-ribosomes; both complexes mediate intranuclear transport of ribosomal precursors |
|
| YDR111C | 1.53 |
ALT2
|
Putative alanine transaminase (glutamic pyruvic transaminase) |
|
| YKL008C | 1.52 |
LAC1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lag1p |
|
| YGR090W | 1.51 |
UTP22
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YLR249W | 1.51 |
YEF3
|
Translational elongation factor 3, stimulates the binding of aminoacyl-tRNA (AA-tRNA) to ribosomes by releasing EF-1 alpha from the ribosomal complex; contains two ABC cassettes; binds and hydrolyses ATP |
|
| YGR027C | 1.49 |
RPS25A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Bp and has similarity to rat S25 ribosomal protein |
|
| YOR222W | 1.48 |
ODC2
|
Mitochondrial inner membrane transporter, exports 2-oxoadipate and 2-oxoglutarate from the mitochondrial matrix to the cytosol for use in lysine and glutamate biosynthesis and in lysine catabolism |
|
| YKL110C | 1.48 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YOR314W | 1.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL265W | 1.45 |
DIP5
|
Dicarboxylic amino acid permease, mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate; also a transporter for Gln, Asn, Ser, Ala, and Gly |
|
| YLR150W | 1.45 |
STM1
|
Protein that binds G4 quadruplex and purine motif triplex nucleic acid; acts with Cdc13p to maintain telomere structure; interacts with ribosomes and subtelomeric Y' DNA; multicopy suppressor of tom1 and pop2 mutations |
|
| YHR152W | 1.45 |
SPO12
|
Nucleolar protein of unknown function, positive regulator of mitotic exit; involved in regulating release of Cdc14p from the nucleolus in early anaphase, may play similar role in meiosis |
|
| YJR016C | 1.44 |
ILV3
|
Dihydroxyacid dehydratase, catalyzes third step in the common pathway leading to biosynthesis of branched-chain amino acids |
|
| YLR254C | 1.44 |
NDL1
|
Homolog of nuclear distribution factor NudE, NUDEL; interacts with Pac1p and regulates dynein targeting to microtubule plus ends |
|
| YBR085W | 1.43 |
AAC3
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; expressed under anaerobic conditions; similar to Pet9p and Aac1p; has roles in maintenance of viability and in respiration |
|
| YBR130C | 1.43 |
SHE3
|
Protein that acts as an adaptor between Myo4p and the She2p-mRNA complex; part of the mRNA localization machinery that restricts accumulation of certain proteins to the bud; also required for cortical ER inheritance |
|
| YPL037C | 1.43 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YLR210W | 1.43 |
CLB4
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the G2/M transition; may be involved in DNA replication and spindle assembly; accumulates during S phase and G2, then targeted for ubiquitin-mediated degradation |
|
| YKL060C | 1.43 |
FBA1
|
Fructose 1,6-bisphosphate aldolase, required for glycolysis and gluconeogenesis; catalyzes conversion of fructose 1,6 bisphosphate to glyceraldehyde-3-P and dihydroxyacetone-P; locates to mitochondrial outer surface upon oxidative stress |
|
| YMR290W-A | 1.43 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YGR106C | 1.42 |
VOA1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar memebrane |
|
| YKL080W | 1.41 |
VMA5
|
Subunit C of the eight-subunit V1 peripheral membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane |
Network of associatons between targets according to the STRING database.
First level regulatory network of ROX1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 2.6 | 7.7 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 1.4 | 2.8 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 1.4 | 5.5 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 1.3 | 9.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 1.2 | 8.7 | GO:0019660 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 1.2 | 7.4 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 1.2 | 4.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 1.1 | 4.6 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 1.1 | 3.3 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 1.1 | 7.5 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.9 | 3.7 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.9 | 3.6 | GO:0090337 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 0.9 | 3.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.9 | 6.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.9 | 7.7 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.8 | 2.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.8 | 3.9 | GO:0007535 | donor selection(GO:0007535) |
| 0.8 | 6.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.7 | 2.9 | GO:0000296 | spermine transport(GO:0000296) |
| 0.7 | 0.7 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.7 | 2.8 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.6 | 0.6 | GO:0035950 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.6 | 1.2 | GO:0016093 | polyprenol metabolic process(GO:0016093) polyprenol biosynthetic process(GO:0016094) dolichol metabolic process(GO:0019348) dolichol biosynthetic process(GO:0019408) |
| 0.6 | 6.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.6 | 1.8 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.6 | 8.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.6 | 1.8 | GO:0042992 | negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.6 | 4.7 | GO:0048309 | endoplasmic reticulum inheritance(GO:0048309) |
| 0.6 | 1.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.5 | 1.1 | GO:0009847 | spore germination(GO:0009847) |
| 0.5 | 2.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.5 | 1.6 | GO:0018216 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.5 | 1.5 | GO:1901351 | regulation of phosphatidylglycerol biosynthetic process(GO:1901351) negative regulation of phosphatidylglycerol biosynthetic process(GO:1901352) |
| 0.5 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.5 | 16.5 | GO:0016126 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.5 | 3.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.5 | 4.9 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.5 | 2.4 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.5 | 1.5 | GO:0043069 | negative regulation of apoptotic process(GO:0043066) negative regulation of programmed cell death(GO:0043069) negative regulation of cell death(GO:0060548) |
| 0.5 | 1.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.5 | 1.4 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.5 | 1.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 2.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.5 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.5 | 7.3 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.5 | 1.8 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.4 | 1.3 | GO:0043902 | starch metabolic process(GO:0005982) starch catabolic process(GO:0005983) regulation of starch catabolic process by regulation of transcription from RNA polymerase II promoter(GO:0035956) positive regulation of starch catabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0035957) positive regulation of multi-organism process(GO:0043902) regulation of starch catabolic process(GO:2000881) positive regulation of starch catabolic process(GO:2000883) regulation of starch metabolic process(GO:2000904) positive regulation of starch metabolic process(GO:2000906) |
| 0.4 | 2.7 | GO:0046513 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.4 | 0.9 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.4 | 0.9 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.4 | 2.6 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.4 | 4.3 | GO:0090662 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.4 | 1.7 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) |
| 0.4 | 3.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.4 | 2.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.4 | 3.3 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.4 | 2.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.4 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 1.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.4 | 1.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.4 | 1.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 3.7 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.4 | 4.5 | GO:0034963 | box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) |
| 0.4 | 1.5 | GO:0000433 | negative regulation of transcription from RNA polymerase II promoter by glucose(GO:0000433) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 1.1 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.4 | 45.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 0.3 | GO:0043067 | regulation of cell death(GO:0010941) regulation of apoptotic process(GO:0042981) regulation of programmed cell death(GO:0043067) |
| 0.3 | 4.5 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.3 | 7.5 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.4 | GO:0019365 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.3 | 4.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.3 | 2.7 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.3 | 1.3 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.9 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.3 | 2.5 | GO:0000321 | re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.3 | 1.8 | GO:0035376 | sterol import(GO:0035376) |
| 0.3 | 1.2 | GO:0036213 | actomyosin contractile ring contraction(GO:0000916) contractile ring contraction(GO:0036213) |
| 0.3 | 1.8 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.3 | 1.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.3 | 1.5 | GO:0060304 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 1.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 2.9 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.3 | 3.4 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.3 | 0.9 | GO:0009394 | pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) 2'-deoxyribonucleotide metabolic process(GO:0009394) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.3 | 1.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 1.1 | GO:0071406 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.3 | 2.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 0.8 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.3 | 1.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 4.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 1.6 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.3 | 1.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.3 | 2.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 7.3 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.3 | 0.8 | GO:2001021 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.2 | 0.7 | GO:0006571 | tyrosine biosynthetic process(GO:0006571) |
| 0.2 | 1.0 | GO:0090220 | meiotic telomere clustering(GO:0045141) establishment of chromosome localization(GO:0051303) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 | 1.7 | GO:0015791 | polyol transport(GO:0015791) |
| 0.2 | 0.7 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 2.9 | GO:0000921 | septin ring assembly(GO:0000921) |
| 0.2 | 0.2 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 0.9 | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay(GO:0070481) |
| 0.2 | 1.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.2 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.2 | 2.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 1.8 | GO:0031506 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.2 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.7 | GO:1900062 | regulation of cell aging(GO:0090342) regulation of replicative cell aging(GO:1900062) |
| 0.2 | 3.1 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.2 | 3.3 | GO:0042797 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.2 | 1.1 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 0.2 | 1.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 0.2 | GO:0045337 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.9 | GO:0071966 | cell wall chitin biosynthetic process(GO:0006038) fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.2 | 0.8 | GO:0007157 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.2 | 0.8 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 0.2 | 0.6 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.2 | 0.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) |
| 0.2 | 5.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 0.6 | GO:0072523 | adenine metabolic process(GO:0046083) nucleobase catabolic process(GO:0046113) purine-containing compound catabolic process(GO:0072523) |
| 0.2 | 1.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 2.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 1.0 | GO:0042435 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.2 | 0.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.2 | 0.2 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.2 | 0.8 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.2 | 0.6 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.2 | 0.4 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 1.3 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.2 | 0.4 | GO:0031384 | regulation of initiation of mating projection growth(GO:0031384) |
| 0.2 | 1.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 0.6 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 1.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 0.7 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 1.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 | 0.9 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.2 | 0.7 | GO:0048279 | vesicle fusion with endoplasmic reticulum(GO:0048279) |
| 0.2 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 3.7 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 2.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.2 | 0.5 | GO:0019413 | acetate biosynthetic process(GO:0019413) |
| 0.2 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 2.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 3.1 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) telomere maintenance via telomere lengthening(GO:0010833) |
| 0.2 | 2.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 1.3 | GO:0006614 | cotranslational protein targeting to membrane(GO:0006613) SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 0.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.3 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 0.3 | GO:0031555 | regulation of DNA-templated transcription, termination(GO:0031554) transcriptional attenuation(GO:0031555) transcription antitermination(GO:0031564) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) |
| 0.1 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.9 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 0.7 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.7 | GO:0001301 | progressive alteration of chromatin involved in cell aging(GO:0001301) |
| 0.1 | 2.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.4 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.1 | 0.8 | GO:0071028 | nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 1.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.5 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) barbed-end actin filament capping(GO:0051016) actin filament capping(GO:0051693) |
| 0.1 | 2.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 3.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.4 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.3 | GO:0010978 | gene silencing involved in chronological cell aging(GO:0010978) gene silencing by RNA(GO:0031047) |
| 0.1 | 0.9 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 2.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.1 | 0.5 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.6 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.1 | 4.6 | GO:0044744 | protein targeting to nucleus(GO:0044744) |
| 0.1 | 14.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 0.4 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.1 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.4 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 0.5 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.1 | 1.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 2.0 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 3.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.1 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.1 | 3.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.1 | 0.6 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.1 | 0.3 | GO:0009088 | threonine biosynthetic process(GO:0009088) |
| 0.1 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.6 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 2.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 2.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.4 | GO:0055075 | cellular potassium ion homeostasis(GO:0030007) potassium ion homeostasis(GO:0055075) |
| 0.1 | 0.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.7 | GO:0044091 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.1 | 0.2 | GO:0032886 | regulation of microtubule-based process(GO:0032886) regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.1 | 0.2 | GO:0009097 | isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.2 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.1 | GO:0043038 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.3 | GO:0008219 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.1 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 1.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:0045833 | negative regulation of lipid metabolic process(GO:0045833) |
| 0.1 | 0.9 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.5 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.1 | 0.5 | GO:0016125 | steroid metabolic process(GO:0008202) sterol metabolic process(GO:0016125) |
| 0.1 | 0.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.2 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.1 | 0.4 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 2.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.1 | 0.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.1 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.9 | GO:0034470 | ncRNA processing(GO:0034470) |
| 0.1 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.1 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.1 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) SNARE complex assembly(GO:0035493) regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.1 | GO:0008299 | isoprenoid metabolic process(GO:0006720) isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 0.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:1901658 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.0 | 0.5 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.0 | 0.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) negative regulation of pseudohyphal growth(GO:2000221) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.3 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.0 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.3 | GO:0005992 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.3 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.1 | GO:0001173 | DNA-templated transcriptional start site selection(GO:0001173) transcriptional start site selection at RNA polymerase II promoter(GO:0001174) |
| 0.0 | 0.2 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.1 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) |
| 0.0 | 1.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.1 | GO:0009231 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.0 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0031023 | microtubule organizing center organization(GO:0031023) spindle pole body organization(GO:0051300) |
| 0.0 | 0.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.5 | 7.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 1.1 | 7.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.1 | 3.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.0 | 4.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.8 | 2.5 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.8 | 3.1 | GO:0044697 | HICS complex(GO:0044697) |
| 0.6 | 4.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.6 | 2.9 | GO:0030689 | Noc complex(GO:0030689) |
| 0.6 | 2.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 2.8 | GO:0031389 | DNA replication factor C complex(GO:0005663) Rad17 RFC-like complex(GO:0031389) Elg1 RFC-like complex(GO:0031391) |
| 0.5 | 3.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 2.1 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.5 | 2.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.5 | 32.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 8.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.5 | 1.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 1.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 3.7 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.5 | 10.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.4 | 2.6 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.4 | 4.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.4 | 42.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.4 | 1.7 | GO:0033179 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.4 | 2.0 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.4 | 2.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.4 | 6.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.4 | 0.4 | GO:0000133 | polarisome(GO:0000133) |
| 0.4 | 2.2 | GO:0032126 | eisosome(GO:0032126) |
| 0.4 | 1.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.4 | 1.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 2.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 1.3 | GO:0034990 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.3 | 1.3 | GO:0032160 | septin filament array(GO:0032160) |
| 0.3 | 1.3 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.3 | 2.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.3 | 1.6 | GO:0032545 | CURI complex(GO:0032545) |
| 0.3 | 3.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 2.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.3 | 0.9 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 1.4 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.3 | 1.1 | GO:0000817 | COMA complex(GO:0000817) |
| 0.3 | 2.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.3 | 0.8 | GO:0043529 | GET complex(GO:0043529) |
| 0.2 | 0.7 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.2 | 0.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 1.8 | GO:0000142 | cellular bud neck contractile ring(GO:0000142) |
| 0.2 | 2.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.4 | GO:0048475 | membrane coat(GO:0030117) clathrin adaptor complex(GO:0030131) coated membrane(GO:0048475) |
| 0.2 | 3.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.8 | GO:0030907 | MBF transcription complex(GO:0030907) |
| 0.2 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 6.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 1.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.9 | GO:0034044 | exomer complex(GO:0034044) |
| 0.2 | 0.5 | GO:0030428 | cell septum(GO:0030428) |
| 0.2 | 8.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.2 | 0.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 0.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.8 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 0.9 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.7 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.4 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.8 | GO:0070772 | PAS complex(GO:0070772) |
| 0.1 | 0.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.6 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 4.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.2 | GO:0042729 | DASH complex(GO:0042729) |
| 0.1 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:0000500 | RNA polymerase I upstream activating factor complex(GO:0000500) |
| 0.1 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 3.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 17.3 | GO:0005933 | cellular bud(GO:0005933) |
| 0.1 | 0.1 | GO:0030286 | dynactin complex(GO:0005869) dynein complex(GO:0030286) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 2.9 | GO:0012506 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 2.6 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 1.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 3.6 | GO:0005816 | microtubule organizing center(GO:0005815) spindle pole body(GO:0005816) |
| 0.1 | 0.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 0.3 | GO:0070823 | HDA1 complex(GO:0070823) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.5 | GO:0030894 | alpha DNA polymerase:primase complex(GO:0005658) replisome(GO:0030894) nuclear replisome(GO:0043601) |
| 0.1 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.2 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 0.2 | GO:0000262 | cytoplasmic chromosome(GO:0000229) mitochondrial chromosome(GO:0000262) |
| 0.1 | 2.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.1 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0033263 | HOPS complex(GO:0030897) CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.1 | GO:0043332 | mating projection tip(GO:0043332) |
| 0.0 | 0.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 5.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.0 | 0.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.0 | 0.1 | GO:0045254 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 2.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 2.4 | 7.1 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 2.0 | 12.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.7 | 6.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 1.6 | 6.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.4 | 4.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 1.4 | 5.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.3 | 4.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 1.3 | 5.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 1.1 | 3.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.1 | 6.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.9 | 3.6 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.9 | 4.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.9 | 3.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.9 | 3.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.8 | 4.8 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.8 | 7.6 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.7 | 3.0 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.7 | 2.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.7 | 10.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.7 | 2.2 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.7 | 2.9 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.7 | 7.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.7 | 3.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.7 | 2.0 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.6 | 14.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.6 | 1.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.6 | 1.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.6 | 2.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.6 | 1.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.6 | 2.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.6 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 1.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.5 | 1.6 | GO:0043812 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.5 | 1.6 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.5 | 2.6 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.5 | 2.4 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.5 | 1.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.5 | 2.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.5 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.4 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.4 | 1.7 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 5.0 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.4 | 1.6 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.4 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.4 | 2.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 6.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.4 | 1.9 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.4 | 1.1 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.4 | 3.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.4 | 2.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.4 | 1.1 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.4 | 1.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.3 | 1.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 1.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.3 | 1.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 1.5 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 12.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.3 | 5.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 0.9 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 0.3 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.3 | 0.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 0.8 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.3 | 55.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 0.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 0.5 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 0.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 2.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.2 | 2.3 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.2 | 0.7 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 1.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.2 | 1.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.2 | 7.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 1.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 0.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.2 | 0.2 | GO:0004805 | trehalose-phosphatase activity(GO:0004805) |
| 0.2 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 1.1 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.2 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 2.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 3.5 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.2 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 1.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 0.9 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.2 | 1.8 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.2 | 6.3 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.2 | 0.9 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.2 | 2.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.2 | 0.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 1.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 0.8 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.2 | 8.8 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.2 | 1.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.2 | 2.2 | GO:0008483 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 0.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 0.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 3.7 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.1 | 4.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.8 | GO:0042123 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.1 | 1.2 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.1 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.4 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 6.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.5 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.3 | GO:0042736 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.1 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 2.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 4.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.6 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.5 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 3.6 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.1 | 0.5 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.1 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 2.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.5 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.3 | GO:0001168 | transcription factor activity, RNA polymerase I upstream control element sequence-specific binding(GO:0001168) |
| 0.1 | 0.6 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 0.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.7 | GO:0019207 | kinase regulator activity(GO:0019207) |
| 0.1 | 1.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.1 | GO:0005525 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.1 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) |
| 0.1 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 0.4 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.1 | 0.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 0.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.3 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 4.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.2 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.0 | 0.2 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.6 | 5.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.4 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 0.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 0.4 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 0.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.9 | 1.8 | REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | Genes involved in The citric acid (TCA) cycle and respiratory electron transport |
| 0.7 | 2.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.5 | 3.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.5 | 1.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 1.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.3 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 6.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.3 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 0.2 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.2 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 1.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 2.5 | REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | Genes involved in Metabolism of lipids and lipoproteins |
| 0.1 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.6 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.1 | 0.5 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.1 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.2 | REACTOME AXON GUIDANCE | Genes involved in Axon guidance |
| 0.0 | 0.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |