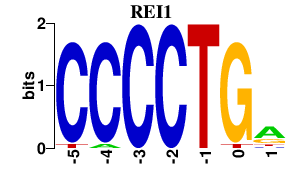
Results for REI1
Z-value: 1.34
Transcription factors associated with REI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REI1
|
S000000471 | Cytoplasmic pre-60S factor |
Activity-expression correlation:
Activity profile of REI1 motif
Sorted Z-values of REI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YDR133C | 10.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YNR001W-A | 9.11 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YFR055W | 8.59 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YFR056C | 8.57 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YPL014W | 5.61 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YGR249W | 5.37 |
MGA1
|
Protein similar to heat shock transcription factor; multicopy suppressor of pseudohyphal growth defects of ammonium permease mutants |
|
| YBR126C | 5.27 |
TPS1
|
Synthase subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; also found in a monomeric form; expression is induced by the stress response and repressed by the Ras-cAMP pathway |
|
| YOL086C | 5.18 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YGL179C | 5.10 |
TOS3
|
Protein kinase, related to and functionally redundant with Elm1p and Sak1p for the phosphorylation and activation of Snf1p; functionally orthologous to LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YIL162W | 4.94 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YOL085C | 4.77 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YBR126W-A | 4.62 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YHL021C | 4.00 |
AIM17
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YPR160W | 3.91 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YBR183W | 3.86 |
YPC1
|
Alkaline ceramidase that also has reverse (CoA-independent) ceramide synthase activity, catalyzes both breakdown and synthesis of phytoceramide; overexpression confers fumonisin B1 resistance |
|
| YKL096W-A | 3.74 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YDR516C | 3.53 |
EMI2
|
Non-essential protein of unknown function required for transcriptional induction of the early meiotic-specific transcription factor IME1; required for sporulation; expression is regulated by glucose-repression transcription factors Mig1/2p |
|
| YER066C-A | 3.48 |
Dubious open reading frame unlikely to encode a protein, partially overlaps uncharacterized ORF YER067W |
||
| YGR138C | 3.45 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YHL003C | 3.38 |
LAG1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lac1p |
|
| YBR182C-A | 3.25 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YNL160W | 3.23 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YOL154W | 3.02 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YGR192C | 3.02 |
TDH3
|
Glyceraldehyde-3-phosphate dehydrogenase, isozyme 3, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3 bis-phosphoglycerate; detected in the cytoplasm and cell-wall |
|
| YGR254W | 2.96 |
ENO1
|
Enolase I, a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and the reverse reaction during gluconeogenesis; expression is repressed in response to glucose |
|
| YJL158C | 2.95 |
CIS3
|
Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
|
| YNL028W | 2.83 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL048C | 2.76 |
UBX6
|
UBX (ubiquitin regulatory X) domain-containing protein that interacts with Cdc48p, transcription is repressed when cells are grown in media containing inositol and choline |
|
| YJR073C | 2.74 |
OPI3
|
Phospholipid methyltransferase (methylene-fatty-acyl-phospholipid synthase), catalyzes the last two steps in phosphatidylcholine biosynthesis |
|
| YGR139W | 2.70 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFL015W-A | 2.69 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFL015C | 2.68 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YBR092C | 2.61 |
PHO3
|
Constitutively expressed acid phosphatase similar to Pho5p; brought to the cell surface by transport vesicles; hydrolyzes thiamin phosphates in the periplasmic space, increasing cellular thiamin uptake; expression is repressed by thiamin |
|
| YHR094C | 2.55 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YER067W | 2.54 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YER067W is not an essential gene |
||
| YDR044W | 2.54 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YDR095C | 2.51 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR258W | 2.49 |
GSY2
|
Glycogen synthase, similar to Gsy1p; expression induced by glucose limitation, nitrogen starvation, heat shock, and stationary phase; activity regulated by cAMP-dependent, Snf1p and Pho85p kinases as well as by the Gac1p-Glc7p phosphatase |
|
| YGR052W | 2.47 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YGL253W | 2.45 |
HXK2
|
Hexokinase isoenzyme 2 that catalyzes phosphorylation of glucose in the cytosol; predominant hexokinase during growth on glucose; functions in the nucleus to repress expression of HXK1 and GLK1 and to induce expression of its own gene |
|
| YPR119W | 2.44 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YDL205C | 2.41 |
HEM3
|
Phorphobilinogen deaminase, catalyzes the conversion of 4-porphobilinogen to hydroxymethylbilane, the third step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus; expression is regulated by Hap2p-Hap3p |
|
| YER066W | 2.32 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YNR014W | 2.26 |
Putative protein of unknown function; expression is cell-cycle regulated, Azf1p-dependent, and heat-inducible |
||
| YMR290C | 2.24 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YDR345C | 2.23 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YDR033W | 2.18 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YFR015C | 2.18 |
GSY1
|
Glycogen synthase with similarity to Gsy2p, the more highly expressed yeast homolog; expression induced by glucose limitation, nitrogen starvation, environmental stress, and entry into stationary phase |
|
| YKL091C | 2.14 |
Putative homolog of Sec14p, which is a phosphatidylinositol/phosphatidylcholine transfer protein involved in lipid metabolism; localizes to the nucleus |
||
| YIL111W | 2.14 |
COX5B
|
Subunit Vb of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during anaerobic growth while its isoform Va (Cox5Ap) is expressed during aerobic growth |
|
| YGR108W | 2.11 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YML103C | 2.07 |
NUP188
|
Subunit of the nuclear pore complex (NPC), involved in the structural organization of the complex and of the nuclear envelope, also involved in nuclear envelope permeability, interacts with Pom152p and Nic96p |
|
| YOR273C | 1.99 |
TPO4
|
Polyamine transport protein, recognizes spermine, putrescine, and spermidine; localizes to the plasma membrane; member of the major facilitator superfamily |
|
| YLR286C | 1.97 |
CTS1
|
Endochitinase, required for cell separation after mitosis; transcriptional activation during late G and early M cell cycle phases is mediated by transcription factor Ace2p |
|
| YER124C | 1.96 |
DSE1
|
Daughter cell-specific protein, may participate in pathways regulating cell wall metabolism; deletion affects cell separation after division and sensitivity to drugs targeted against the cell wall |
|
| YLR150W | 1.96 |
STM1
|
Protein that binds G4 quadruplex and purine motif triplex nucleic acid; acts with Cdc13p to maintain telomere structure; interacts with ribosomes and subtelomeric Y' DNA; multicopy suppressor of tom1 and pop2 mutations |
|
| YPR074C | 1.96 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YLR109W | 1.94 |
AHP1
|
Thiol-specific peroxiredoxin, reduces hydroperoxides to protect against oxidative damage; function in vivo requires covalent conjugation to Urm1p |
|
| YNL178W | 1.94 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YMR290W-A | 1.93 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YJL153C | 1.91 |
INO1
|
Inositol 1-phosphate synthase, involved in synthesis of inositol phosphates and inositol-containing phospholipids; transcription is coregulated with other phospholipid biosynthetic genes by Ino2p and Ino4p, which bind the UASINO DNA element |
|
| YER131W | 1.85 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YMR261C | 1.84 |
TPS3
|
Regulatory subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; expression is induced by stress conditions and repressed by the Ras-cAMP pathway |
|
| YNL300W | 1.83 |
Glycosylphosphatidylinositol-dependent cell wall protein, expression is periodic and decreases in respone to ergosterol perturbation or upon entry into stationary phase; depletion increases resistance to lactic acid |
||
| YDR517W | 1.83 |
GRH1
|
Acetylated, cis-golgi localized protein involved in ER to Golgi transport; homolog of human GRASP65; forms a complex with the coiled-coil protein Bug1p; mutants are compromised for the fusion of ER-derived vesicles with Golgi membranes |
|
| YLR177W | 1.81 |
Putative protein of unknown function; phosphorylated by Dbf2p-Mob1p in vitro; some strains contain microsatellite polymophisms at this locus; YLR177W is not an essential gene |
||
| YDL055C | 1.80 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YPL037C | 1.75 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YGR234W | 1.75 |
YHB1
|
Nitric oxide oxidoreductase, flavohemoglobin involved in nitric oxide detoxification; plays a role in the oxidative and nitrosative stress responses |
|
| YPR157W | 1.73 |
Putative protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
||
| YGR086C | 1.73 |
PIL1
|
Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways; detected in phosphorylated state in mitochondria |
|
| YJR094W-A | 1.71 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YBR206W | 1.70 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene KTR3 |
||
| YGL202W | 1.69 |
ARO8
|
Aromatic aminotransferase I, expression is regulated by general control of amino acid biosynthesis |
|
| YHR180W-A | 1.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YML052W | 1.65 |
SUR7
|
Putative integral membrane protein; component of eisosomes; associated with endocytosis, along with Pil1p and Lsp1p; sporulation and plasma membrane sphingolipid content are altered in mutants |
|
| YDL023C | 1.63 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YBR158W | 1.63 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YKL008C | 1.63 |
LAC1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lag1p |
|
| YOL101C | 1.61 |
IZH4
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by fatty acids and altered zinc levels; deletion reduces sensitivity to excess zinc; possible role in sterol metabolism |
|
| YJL196C | 1.58 |
ELO1
|
Elongase I, medium-chain acyl elongase, catalyzes carboxy-terminal elongation of unsaturated C12-C16 fatty acyl-CoAs to C16-C18 fatty acids |
|
| YEL001C | 1.57 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YPR149W | 1.56 |
NCE102
|
Protein of unknown function; contains transmembrane domains; involved in secretion of proteins that lack classical secretory signal sequences; component of the detergent-insoluble glycolipid-enriched complexes (DIGs) |
|
| YMR266W | 1.55 |
RSN1
|
Membrane protein of unknown function; overexpression suppresses NaCl sensitivity of sro7 mutant |
|
| YHR181W | 1.55 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YLR154C | 1.53 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YNL066W | 1.53 |
SUN4
|
Cell wall protein related to glucanases, possibly involved in cell wall septation; member of the SUN family |
|
| YPR196W | 1.52 |
Putative maltose activator |
||
| YMR297W | 1.52 |
PRC1
|
Vacuolar carboxypeptidase Y (proteinase C), broad-specificity C-terminal exopeptidase involved in non-specific protein degradation in the vacuole; member of the serine carboxypeptidase family |
|
| YER001W | 1.49 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YER088C-A | 1.49 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YMR082C | 1.48 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL012W | 1.48 |
ERG4
|
C-24(28) sterol reductase, catalyzes the final step in ergosterol biosynthesis; mutants are viable, but lack ergosterol |
|
| YLR257W | 1.47 |
Putative protein of unknown function |
||
| YCR009C | 1.42 |
RVS161
|
Amphiphysin-like lipid raft protein; subunit of a complex (Rvs161p-Rvs167p) that regulates polarization of the actin cytoskeleton, endocytosis, cell polarity, cell fusion and viability following starvation or osmotic stress |
|
| YDR541C | 1.42 |
Putative dihydrokaempferol 4-reductase |
||
| YJR103W | 1.42 |
URA8
|
Minor CTP synthase isozyme (see also URA7), catalyzes the ATP-dependent transfer of the amide nitrogen from glutamine to UTP, forming CTP, the final step in de novo biosynthesis of pyrimidines; involved in phospholipid biosynthesis |
|
| YDL022W | 1.42 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YMR262W | 1.41 |
Protein of unknown function; interacts weakly with Knr4p; YMR262W is not an essential gene |
||
| YOR317W | 1.40 |
FAA1
|
Long chain fatty acyl-CoA synthetase with a preference for C12:0-C16:0 fatty acids; involved in the activation of imported fatty acids; localized to both lipid particles and mitochondrial outer membrane; essential for stationary phase |
|
| YEL056W | 1.40 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YHR163W | 1.37 |
SOL3
|
6-phosphogluconolactonase, catalyzes the second step of the pentose phosphate pathway; weak multicopy suppressor of los1-1 mutation; homologous to Sol2p and Sol1p |
|
| YDR185C | 1.37 |
Mitochondrial protein of unknown function; has similarity to Ups1p, which is involved in regulation of alternative topogenesis of the dynamin-related GTPase Mgm1p |
||
| YDR344C | 1.37 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAL033W | 1.37 |
POP5
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YJR074W | 1.36 |
MOG1
|
Conserved nuclear protein that interacts with GTP-Gsp1p, which is a Ran homolog of the Ras GTPase family, and stimulates nucleotide release, involved in nuclear protein import, nucleotide release is inhibited by Yrb1p |
|
| YPL247C | 1.35 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; similar to the petunia WD repeat protein an11; YPL247C is not an essential gene |
||
| YGR279C | 1.34 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YNL029C | 1.34 |
KTR5
|
Putative mannosyltransferase involved in protein glycosylation; member of the KRE2/MNT1 mannosyltransferase family |
|
| YBL044W | 1.34 |
Putative protein of unknown function; YBL044W is not an essential protein |
||
| YKL182W | 1.33 |
FAS1
|
Beta subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids; contains acetyltransacylase, dehydratase, enoyl reductase, malonyl transacylase, and palmitoyl transacylase activities |
|
| YER130C | 1.33 |
Hypothetical protein |
||
| YKL090W | 1.29 |
CUE2
|
Protein of unknown function; has two CUE domains that bind ubiquitin, which may facilitate intramolecular monoubiquitination |
|
| YOR226C | 1.26 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YDR309C | 1.23 |
GIC2
|
Redundant rho-like GTPase Cdc42p effector; homolog of Gic1p; involved in initiation of budding and cellular polarization; interacts with Cdc42p via the Cdc42/Rac-interactive binding (CRIB) domain and with PI(4,5)P2 via a polybasic region |
|
| YGL148W | 1.23 |
ARO2
|
Bifunctional chorismate synthase and flavin reductase, catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate (EPSP) to form chorismate, which is a precursor to aromatic amino acids |
|
| YIL051C | 1.21 |
MMF1
|
Mitochondrial protein involved in maintenance of the mitochondrial genome |
|
| YHR162W | 1.21 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion |
||
| YER083C | 1.21 |
GET2
|
Subunit of the GET complex; required for meiotic nuclear division and for the retrieval of HDEL proteins from the Golgi to the ER in an ERD2 dependent fashion; may be involved in cell wall function |
|
| YGL199C | 1.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YIP4/YGL198W |
||
| YHR010W | 1.20 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YBR085C-A | 1.20 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YLR437C | 1.19 |
Putative protein of unknown function; epitope tagged protein localizes to the cytoplasm |
||
| YDR276C | 1.19 |
PMP3
|
Small plasma membrane protein related to a family of plant polypeptides that are overexpressed under high salt concentration or low temperature, not essential for viability, deletion causes hyperpolarization of the plasma membrane potential |
|
| YGL225W | 1.19 |
VRG4
|
Golgi GDP-mannose transporter; regulates Golgi function and glycosylation in Golgi |
|
| YJR113C | 1.19 |
RSM7
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S7 ribosomal protein |
|
| YNL193W | 1.18 |
Putative protein of unknown function; exhibits a two-hybrid interaction with Yhr151cp in a large-scale analysis |
||
| YPL263C | 1.17 |
KEL3
|
Cytoplasmic protein of unknown function |
|
| YFR054C | 1.17 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR094W | 1.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF DNF2 |
||
| YLL050C | 1.14 |
COF1
|
Cofilin, promotes actin filament depolarization in a pH-dependent manner; binds both actin monomers and filaments and severs filaments; thought to be regulated by phosphorylation at SER4; ubiquitous and essential in eukaryotes |
|
| YJR102C | 1.14 |
VPS25
|
Component of the ESCRT-II complex, which is involved in ubiquitin-dependent sorting of proteins into the endosome |
|
| YPL274W | 1.14 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YDL022C-A | 1.14 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene DIA3; identified by fungal homology and RT-PCR |
||
| YMR205C | 1.13 |
PFK2
|
Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes |
|
| YKL128C | 1.13 |
PMU1
|
Putative phosphomutase, contains a region homologous to the active site of phosphomutases; overexpression suppresses the histidine auxotrophy of an ade3 ade16 ade17 triple mutant and the temperature sensitivity of a tps2 mutant |
|
| YHL028W | 1.13 |
WSC4
|
ER membrane protein involved in the translocation of soluble secretory proteins and insertion of membrane proteins into the ER membrane; may also have a role in the stress response but has only partial functional overlap with WSC1-3 |
|
| YGR127W | 1.13 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p, indicating a possible role in stress response |
||
| YKR074W | 1.12 |
AIM29
|
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YHR190W | 1.12 |
ERG9
|
Farnesyl-diphosphate farnesyl transferase (squalene synthase), joins two farnesyl pyrophosphate moieties to form squalene in the sterol biosynthesis pathway |
|
| YPL087W | 1.10 |
YDC1
|
Alkaline dihydroceramidase, involved in sphingolipid metabolism; preferentially hydrolyzes dihydroceramide to a free fatty acid and dihydrosphingosine; has a minor reverse activity |
|
| YLR154W-B | 1.10 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YEL073C | 1.10 |
Putative protein of unknown function; located adjacent to ARS503 and the telomere on the left arm of chromosome V; regulated by inositol/choline |
||
| YDR279W | 1.08 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YBR177C | 1.08 |
EHT1
|
Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis; possesses short-chain esterase activity; localizes to lipid particles and the mitochondrial outer membrane |
|
| YDR074W | 1.07 |
TPS2
|
Phosphatase subunit of the trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; expression is induced by stress conditions and repressed by the Ras-cAMP pathway |
|
| YER177W | 1.07 |
BMH1
|
14-3-3 protein, major isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YOL155C | 1.06 |
HPF1
|
Haze-protective mannoprotein that reduces the particle size of aggregated proteins in white wines |
|
| YGR242W | 1.05 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YAP1802/YGR241C |
||
| YGR107W | 1.04 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR241C | 1.03 |
YAP1802
|
Protein involved in clathrin cage assembly; binds Pan1p and clathrin; homologous to Yap1801p, member of the AP180 protein family |
|
| YEL038W | 1.03 |
UTR4
|
Protein of unknown function with sequence similarity to 2,3-diketo-5-methylthiopentyl-1-phosphate enolase-phosphatases, found in both the cytoplasm and nucleus |
|
| YDL125C | 1.02 |
HNT1
|
Adenosine 5'-monophosphoramidase; interacts physically and genetically with Kin28p, a CDK and TFIIK subunit, and genetically with CAK1; member of the histidine triad (HIT) superfamily of nucleotide-binding proteins and similar to Hint |
|
| YPR063C | 1.02 |
ER-localized protein of unknown function |
||
| YAL038W | 1.02 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YHR141C | 1.00 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YMR246W | 1.00 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YGR020C | 1.00 |
VMA7
|
Subunit F of the eight-subunit V1 peripheral membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic proton pump found throughout the endomembrane system; required for the V1 domain to assemble onto the vacuolar membrane |
|
| YGR050C | 0.99 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGL037C | 0.98 |
PNC1
|
Nicotinamidase that converts nicotinamide to nicotinic acid as part of the NAD(+) salvage pathway, required for life span extension by calorie restriction; PNC1 expression responds to all known stimuli that extend replicative life span |
|
| YJL007C | 0.98 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNR013C | 0.98 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YLR312W-A | 0.97 |
MRPL15
|
Mitochondrial ribosomal protein of the large subunit |
|
| YJR114W | 0.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSM7/YJR113C |
||
| YFL022C | 0.96 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YKR066C | 0.95 |
CCP1
|
Mitochondrial cytochrome-c peroxidase; degrades reactive oxygen species in mitochondria, involved in the response to oxidative stress |
|
| YML004C | 0.95 |
GLO1
|
Monomeric glyoxalase I, catalyzes the detoxification of methylglyoxal (a by-product of glycolysis) via condensation with glutathione to produce S-D-lactoylglutathione; expression regulated by methylglyoxal levels and osmotic stress |
|
| YDR245W | 0.94 |
MNN10
|
Subunit of a Golgi mannosyltransferase complex also containing Anp1p, Mnn9p, Mnn11p, and Hoc1p that mediates elongation of the polysaccharide mannan backbone; membrane protein of the mannosyltransferase family |
|
| YOR013W | 0.94 |
IRC11
|
Dubious opening reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YOR012C; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YNL030W | 0.93 |
HHF2
|
One of two identical histone H4 proteins (see also HHF1); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YDR002W | 0.93 |
YRB1
|
Ran GTPase binding protein; involved in nuclear protein import and RNA export, ubiquitin-mediated protein degradation during the cell cycle; shuttles between the nucleus and cytoplasm; is essential; homolog of human RanBP1 |
|
| YOR320C | 0.93 |
GNT1
|
N-acetylglucosaminyltransferase capable of modification of N-linked glycans in the Golgi apparatus |
|
| YGR131W | 0.93 |
Protein of unknown function; expression induced in response to ketoconazole; promoter region contains a sterol regulatory element motif, which has been identified as a Upc2p-binding site |
||
| YDR385W | 0.93 |
EFT2
|
Elongation factor 2 (EF-2), also encoded by EFT1; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YHR025W | 0.90 |
THR1
|
Homoserine kinase, conserved protein required for threonine biosynthesis; expression is regulated by the GCN4-mediated general amino acid control pathway |
|
| YPR148C | 0.90 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YER182W | 0.90 |
FMP10
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL301C | 0.90 |
RPL18B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Ap and has similarity to rat L18 ribosomal protein |
|
| YML102W | 0.90 |
CAC2
|
Component of the chromatin assembly complex (with Rlf2p and Msi1p) that assembles newly synthesized histones onto recently replicated DNA, required for building functional kinetochores, conserved from yeast to humans |
|
| YLR012C | 0.89 |
Putative protein of unknown function; YLR012C is not an essential gene |
||
| YLR029C | 0.89 |
RPL15A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Bp and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YLR206W | 0.89 |
ENT2
|
Epsin-like protein required for endocytosis and actin patch assembly and functionally redundant with Ent1p; contains clathrin-binding motif at C-terminus |
|
| YDR381W | 0.89 |
YRA1
|
Nuclear protein that binds to RNA and to Mex67p, required for export of poly(A)+ mRNA from the nucleus; member of the REF (RNA and export factor binding proteins) family; another family member, Yra2p, can substitute for Yra1p function |
|
| YGL040C | 0.88 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YMR194C-A | 0.87 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER019C-A | 0.87 |
SBH2
|
Ssh1p-Sss1p-Sbh2p complex component, involved in protein translocation into the endoplasmic reticulum; homologous to Sbh1p |
|
| YGR264C | 0.86 |
MES1
|
Methionyl-tRNA synthetase, forms a complex with glutamyl-tRNA synthetase (Gus1p) and Arc1p, which increases the catalytic efficiency of both tRNA synthetases; also has a role in nuclear export of tRNAs |
|
| YMR083W | 0.86 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YOL030W | 0.85 |
GAS5
|
1,3-beta-glucanosyltransferase, has similarity to Gas1p; localizes to the cell wall |
|
| YOR376W-A | 0.84 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YOR047C | 0.83 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YGL082W | 0.82 |
Putative protein of unknown function; predicted prenylation/proteolysis target of Afc1p and Rce1p; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YGL082W is not an essential gene |
||
| YAL029C | 0.82 |
MYO4
|
One of two type V myosin motors (along with MYO2) involved in actin-based transport of cargos; required for mRNA transport, including ASH1 mRNA, and facilitating the growth and movement of ER tubules into the growing bud along with She3p |
|
| YCR031C | 0.81 |
RPS14A
|
Ribosomal protein 59 of the small subunit, required for ribosome assembly and 20S pre-rRNA processing; mutations confer cryptopleurine resistance; nearly identical to Rps14Bp and similar to E. coli S11 and rat S14 ribosomal proteins |
|
| YKL127W | 0.80 |
PGM1
|
Phosphoglucomutase, minor isoform; catalyzes the conversion from glucose-1-phosphate to glucose-6-phosphate, which is a key step in hexose metabolism |
|
| YPL015C | 0.79 |
HST2
|
Cytoplasmic member of the silencing information regulator 2 (Sir2) family of NAD(+)-dependent protein deacetylases; modulates nucleolar (rDNA) and telomeric silencing; possesses NAD(+)-dependent histone deacetylase activity in vitro |
|
| YGR266W | 0.78 |
Protein of unknown function, predicted to contain a single transmembrane domain; localized to both the mitochondrial outer membrane and the plasma membrane |
||
| YJR009C | 0.78 |
TDH2
|
Glyceraldehyde-3-phosphate dehydrogenase, isozyme 2, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3 bis-phosphoglycerate; detected in the cytoplasm and cell-wall |
|
| YDL101C | 0.78 |
DUN1
|
Cell-cycle checkpoint serine-threonine kinase required for DNA damage-induced transcription of certain target genes, phosphorylation of Rad55p and Sml1p, and transient G2/M arrest after DNA damage; also regulates postreplicative DNA repair |
|
| YOR293W | 0.77 |
RPS10A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps10Bp and has similarity to rat ribosomal protein S10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of REI1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 1.7 | 5.0 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 1.5 | 6.2 | GO:0000296 | spermine transport(GO:0000296) |
| 1.3 | 9.2 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 1.3 | 5.1 | GO:0046513 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.9 | 6.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.7 | 5.1 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.7 | 2.0 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.7 | 4.6 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.6 | 2.4 | GO:0035337 | long-chain fatty acid transport(GO:0015909) fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.6 | 1.8 | GO:0060548 | negative regulation of apoptotic process(GO:0043066) negative regulation of programmed cell death(GO:0043069) negative regulation of cell death(GO:0060548) |
| 0.6 | 1.1 | GO:0015848 | spermidine transport(GO:0015848) |
| 0.6 | 1.7 | GO:1902223 | tyrosine biosynthetic process(GO:0006571) L-phenylalanine biosynthetic process(GO:0009094) erythrose 4-phosphate/phosphoenolpyruvate family amino acid biosynthetic process(GO:1902223) |
| 0.6 | 2.2 | GO:0046323 | mannose metabolic process(GO:0006013) glucose import(GO:0046323) |
| 0.5 | 4.2 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.5 | 2.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.5 | 2.9 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.5 | 1.4 | GO:0065005 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.5 | 11.8 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.5 | 1.8 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.4 | 1.8 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.4 | 1.3 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.4 | 1.7 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.4 | 2.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.4 | 2.0 | GO:1901072 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.4 | 1.9 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.4 | 1.9 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.4 | 2.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.9 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.4 | 2.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.3 | 1.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.3 | 2.0 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 5.9 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.3 | 1.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 0.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.3 | 1.2 | GO:0046417 | chorismate biosynthetic process(GO:0009423) chorismate metabolic process(GO:0046417) |
| 0.3 | 1.4 | GO:0009208 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.3 | 0.8 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.3 | 0.8 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.3 | 1.6 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.3 | 1.8 | GO:0010969 | regulation of pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0010969) regulation of signal transduction involved in conjugation with cellular fusion(GO:0060238) |
| 0.3 | 0.8 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 3.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 2.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.7 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.2 | 1.2 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.2 | 0.5 | GO:0043097 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.2 | 1.0 | GO:0019357 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.2 | 0.6 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.2 | 1.7 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 0.2 | 2.3 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.2 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 1.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.2 | 3.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.9 | GO:0045338 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.7 | GO:0008299 | isoprenoid metabolic process(GO:0006720) isoprenoid biosynthetic process(GO:0008299) |
| 0.2 | 0.4 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.2 | 1.8 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.2 | 1.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 4.4 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.2 | 1.8 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.2 | 1.4 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.2 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.2 | 3.6 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.2 | 0.5 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.1 | 1.3 | GO:0034965 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.1 | 0.4 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) |
| 0.1 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 1.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.0 | GO:0048309 | endoplasmic reticulum inheritance(GO:0048309) |
| 0.1 | 0.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.9 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 0.5 | GO:0098610 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.1 | 0.3 | GO:0016320 | endoplasmic reticulum membrane fusion(GO:0016320) |
| 0.1 | 0.6 | GO:1901070 | GMP biosynthetic process(GO:0006177) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.1 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.7 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 1.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 0.2 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 0.9 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.4 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.1 | 1.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.3 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.1 | 0.3 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.1 | 0.4 | GO:1900434 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.1 | 1.7 | GO:0044108 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.1 | 0.3 | GO:0018216 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.2 | GO:0072369 | regulation of lipid transport(GO:0032368) positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 1.2 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.5 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.7 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.1 | 2.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.1 | 0.2 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.1 | 2.5 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.1 | 0.2 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 0.3 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 0.3 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.1 | 0.2 | GO:0046083 | adenine metabolic process(GO:0046083) |
| 0.1 | 9.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.2 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.1 | 0.3 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.1 | 0.2 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.1 | 0.5 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.1 | 0.4 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 | 0.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.6 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 2.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.2 | GO:0042797 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.5 | GO:0051278 | fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.4 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.1 | 0.7 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.5 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.1 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.4 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.1 | 0.2 | GO:1900542 | regulation of purine nucleotide metabolic process(GO:1900542) |
| 0.1 | 0.7 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 1.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.1 | GO:0009409 | response to cold(GO:0009409) cellular response to cold(GO:0070417) |
| 0.1 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.0 | 0.1 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 1.6 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.0 | GO:0032956 | regulation of actin cytoskeleton organization(GO:0032956) regulation of actin filament-based process(GO:0032970) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0007535 | donor selection(GO:0007535) |
| 0.0 | 0.7 | GO:0042723 | thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.6 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:1901661 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) ketone biosynthetic process(GO:0042181) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.3 | GO:0032120 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.1 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0043173 | purine nucleotide salvage(GO:0032261) purine-containing compound salvage(GO:0043101) nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0030541 | plasmid partitioning(GO:0030541) |
| 0.0 | 0.6 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.4 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.1 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0040001 | establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) establishment of spindle orientation(GO:0051294) |
| 0.0 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0015940 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0010695 | regulation of spindle pole body separation(GO:0010695) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 1.1 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.1 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.0 | 0.1 | GO:0000161 | MAPK cascade involved in osmosensory signaling pathway(GO:0000161) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0051598 | meiotic cell cycle checkpoint(GO:0033313) meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 2.4 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.0 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 1.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0048024 | regulation of RNA splicing(GO:0043484) regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.0 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.0 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.1 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.0 | 0.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.0 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) microtubule polymerization(GO:0046785) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.9 | 2.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.6 | 1.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 2.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 1.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 1.3 | GO:0043529 | GET complex(GO:0043529) |
| 0.4 | 2.4 | GO:0032126 | eisosome(GO:0032126) |
| 0.4 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.4 | 1.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 1.4 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.3 | 1.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 2.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 0.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 22.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.3 | 0.9 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 0.3 | 0.9 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.3 | 0.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 3.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.3 | 1.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 6.5 | GO:0009277 | fungal-type cell wall(GO:0009277) |
| 0.2 | 2.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.2 | 0.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 1.7 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.2 | 2.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 0.5 | GO:0000930 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.9 | GO:0000136 | alpha-1,6-mannosyltransferase complex(GO:0000136) |
| 0.2 | 0.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 0.9 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.1 | 1.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 11.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.9 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 2.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 7.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 1.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 8.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 3.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.3 | GO:0044697 | HICS complex(GO:0044697) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.8 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 1.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) tubulin complex(GO:0045298) |
| 0.1 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.3 | GO:0045254 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.2 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 0.1 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 11.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 0.4 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 3.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0034456 | UTP-C complex(GO:0034456) |
| 0.0 | 0.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.7 | GO:0005815 | microtubule organizing center(GO:0005815) spindle pole body(GO:0005816) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 0.1 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.1 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.7 | GO:0030479 | actin cortical patch(GO:0030479) endocytic patch(GO:0061645) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.0 | GO:0030907 | MBF transcription complex(GO:0030907) SBF transcription complex(GO:0033309) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 2.2 | 9.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.6 | 4.9 | GO:0004564 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 1.5 | 6.2 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 1.5 | 9.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.4 | 5.8 | GO:0004396 | hexokinase activity(GO:0004396) |
| 1.3 | 3.8 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.1 | 6.5 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.1 | 1.1 | GO:0004805 | trehalose-phosphatase activity(GO:0004805) |
| 1.0 | 3.9 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.7 | 2.7 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.7 | 2.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.6 | 1.9 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.6 | 2.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.6 | 2.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.6 | 3.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.6 | 1.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.6 | 1.8 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.6 | 1.1 | GO:0015606 | spermidine transmembrane transporter activity(GO:0015606) |
| 0.5 | 6.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.5 | 4.3 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.5 | 1.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.4 | 2.0 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.4 | 1.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.4 | 1.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.4 | 1.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 2.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.3 | 3.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 1.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.3 | 1.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 1.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.3 | 1.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 1.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 0.9 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.3 | 1.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 1.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 2.6 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.3 | 1.3 | GO:0004026 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.2 | 1.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 1.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 1.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 2.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 1.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 0.7 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.2 | 1.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 1.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.2 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 1.1 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.2 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.9 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 1.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.8 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 2.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.8 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.1 | 0.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 3.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.7 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.1 | 3.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.7 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.9 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 1.0 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.1 | 20.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.7 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.1 | 2.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.6 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.2 | GO:0016972 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) thiol oxidase activity(GO:0016972) |
| 0.1 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.3 | GO:0016725 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0015188 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.1 | 1.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 2.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 1.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.5 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.2 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.1 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0015151 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 1.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0035591 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.2 | GO:0099516 | ion antiporter activity(GO:0099516) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.3 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.0 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 36.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.0 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.7 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.7 | 2.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.4 | 1.4 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.3 | 2.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 3.8 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.2 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 0.6 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.1 | 0.4 | REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | Genes involved in Cytokine Signaling in Immune system |
| 0.1 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.2 | REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | Genes involved in Transmembrane transport of small molecules |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 38.5 | REACTOME GLYCOSAMINOGLYCAN METABOLISM | Genes involved in Glycosaminoglycan metabolism |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME REGULATION OF MITOTIC CELL CYCLE | Genes involved in Regulation of mitotic cell cycle |
| 0.0 | 0.3 | REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | Genes involved in Metabolism of lipids and lipoproteins |
| 0.0 | 0.0 | REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | Genes involved in The citric acid (TCA) cycle and respiratory electron transport |