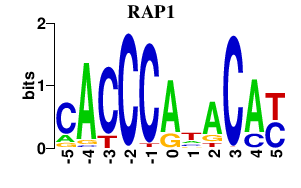
Results for RAP1
Z-value: 8.56
Transcription factors associated with RAP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RAP1
|
S000005160 | Essential DNA-binding transcription regulator that binds at many loci |
Activity-expression correlation:
Activity profile of RAP1 motif
Sorted Z-values of RAP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YJR094W-A | 98.78 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YNL338W | 82.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps TEL14L-XC, which is Telomeric X element Core sequence on the left arm of Chromosome XIV |
||
| YER117W | 70.57 |
RPL23B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Ap and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YFR055W | 69.31 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YLR167W | 65.79 |
RPS31
|
Fusion protein that is cleaved to yield a ribosomal protein of the small (40S) subunit and ubiquitin; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes; interacts genetically with translation factor eIF2B |
|
| YPL250W-A | 65.11 |
Identified by fungal homology and RT-PCR |
||
| YOR293W | 64.33 |
RPS10A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps10Bp and has similarity to rat ribosomal protein S10 |
|
| YHL015W | 63.89 |
RPS20
|
Protein component of the small (40S) ribosomal subunit; overproduction suppresses mutations affecting RNA polymerase III-dependent transcription; has similarity to E. coli S10 and rat S20 ribosomal proteins |
|
| YMR142C | 63.24 |
RPL13B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Ap; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YPL079W | 63.12 |
RPL21B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl21Ap and has similarity to rat L21 ribosomal protein |
|
| YMR143W | 62.45 |
RPS16A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Bp and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YIL018W | 62.04 |
RPL2B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Ap and has similarity to E. coli L2 and rat L8 ribosomal proteins; expression is upregulated at low temperatures |
|
| YFR056C | 61.42 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YPL090C | 60.33 |
RPS6A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Bp and has similarity to rat S6 ribosomal protein |
|
| YDR471W | 60.23 |
RPL27B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Ap and has similarity to rat L27 ribosomal protein |
|
| YOR096W | 58.94 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YDL082W | 57.68 |
RPL13A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Bp; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YGR034W | 56.78 |
RPL26B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Ap and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YGL103W | 56.25 |
RPL28
|
Ribosomal protein of the large (60S) ribosomal subunit, has similarity to E. coli L15 and rat L27a ribosomal proteins; may have peptidyl transferase activity; can mutate to cycloheximide resistance |
|
| YJL177W | 55.73 |
RPL17B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl17Ap and has similarity to E. coli L22 and rat L17 ribosomal proteins |
|
| YGR148C | 55.70 |
RPL24B
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Ap and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YHR141C | 55.21 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YDL083C | 55.01 |
RPS16B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Ap and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YBR181C | 54.69 |
RPS6B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps6Ap and has similarity to rat S6 ribosomal protein |
|
| YNL337W | 54.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJL190C | 54.10 |
RPS22A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Bp and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YGR214W | 52.39 |
RPS0A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Bp; required for maturation of 18S rRNA along with Rps0Bp; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YLR448W | 52.17 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YBR048W | 51.24 |
RPS11B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Ap and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YDR033W | 51.00 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YER131W | 50.80 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YOL127W | 50.67 |
RPL25
|
Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit, has similarity to E. coli L23 and rat L23a ribosomal proteins; binds to 26S rRNA via a conserved C-terminal motif |
|
| YJL136C | 50.28 |
RPS21B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Ap and has similarity to rat S21 ribosomal protein |
|
| YDR447C | 50.08 |
RPS17B
|
Ribosomal protein 51 (rp51) of the small (40s) subunit; nearly identical to Rps17Ap and has similarity to rat S17 ribosomal protein |
|
| YEL054C | 49.51 |
RPL12A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Bp; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YDR418W | 48.68 |
RPL12B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Ap; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YGL135W | 48.01 |
RPL1B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl1Ap and has similarity to E. coli L1 and rat L10a ribosomal proteins; rpl1a rpl1b double null mutation is lethal |
|
| YKR094C | 47.39 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YER056C-A | 47.28 |
RPL34A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Bp and has similarity to rat L34 ribosomal protein |
|
| YJR145C | 46.33 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YML073C | 46.32 |
RPL6A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Bp and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YNL069C | 45.96 |
RPL16B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Ap, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YOL121C | 45.83 |
RPS19A
|
Protein component of the small (40S) ribosomal subunit, required for assembly and maturation of pre-40 S particles; mutations in human RPS19 are associated with Diamond Blackfan anemia; nearly identical to Rps19Bp |
|
| YML024W | 45.53 |
RPS17A
|
Ribosomal protein 51 (rp51) of the small (40s) subunit; nearly identical to Rps17Bp and has similarity to rat S17 ribosomal protein |
|
| YFR031C-A | 45.08 |
RPL2A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Bp and has similarity to E. coli L2 and rat L8 ribosomal proteins |
|
| YOL086C | 44.86 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YBR189W | 44.06 |
RPS9B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps9Ap and has similarity to E. coli S4 and rat S9 ribosomal proteins |
|
| YDR064W | 43.71 |
RPS13
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S15 and rat S13 ribosomal proteins |
|
| YGL076C | 43.60 |
RPL7A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl7Bp and has similarity to E. coli L30 and rat L7 ribosomal proteins; contains a conserved C-terminal Nucleic acid Binding Domain (NDB2) |
|
| YBL092W | 43.07 |
RPL32
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L32 ribosomal protein; overexpression disrupts telomeric silencing |
|
| YLR441C | 42.92 |
RPS1A
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Bp and has similarity to rat S3a ribosomal protein |
|
| YDR025W | 42.91 |
RPS11A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Bp and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YIL052C | 42.52 |
RPL34B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Ap and has similarity to rat L34 ribosomal protein |
|
| YGR027C | 41.71 |
RPS25A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Bp and has similarity to rat S25 ribosomal protein |
|
| YPR132W | 41.33 |
RPS23B
|
Ribosomal protein 28 (rp28) of the small (40S) ribosomal subunit, required for translational accuracy; nearly identical to Rps23Ap and similar to E. coli S12 and rat S23 ribosomal proteins; deletion of both RPS23A and RPS23B is lethal |
|
| YPR102C | 41.01 |
RPL11A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl11Bp; involved in ribosomal assembly; depletion causes degradation of proteins and RNA of the 60S subunit; has similarity to E. coli L5 and rat L11 |
|
| YGL189C | 40.95 |
RPS26A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Bp and has similarity to rat S26 ribosomal protein |
|
| YML063W | 40.76 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YGR118W | 40.73 |
RPS23A
|
Ribosomal protein 28 (rp28) of the small (40S) ribosomal subunit, required for translational accuracy; nearly identical to Rps23Bp and similar to E. coli S12 and rat S23 ribosomal proteins; deletion of both RPS23A and RPS23B is lethal |
|
| YLR061W | 40.30 |
RPL22A
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl22Bp and to rat L22 ribosomal protein |
|
| YPL037C | 39.96 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YML026C | 39.57 |
RPS18B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Ap and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YBR118W | 38.75 |
TEF2
|
Translational elongation factor EF-1 alpha; also encoded by TEF1; functions in the binding reaction of aminoacyl-tRNA (AA-tRNA) to ribosomes |
|
| YOR234C | 38.12 |
RPL33B
|
Ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Ap and has similarity to rat L35a; rpl33b null mutant exhibits normal growth while rpl33a rpl33b double null mutant is inviable |
|
| YBR084C-A | 38.06 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YLR154C | 37.54 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YGL123C-A | 36.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPS2/YGL123W |
||
| YGL123W | 36.58 |
RPS2
|
Protein component of the small (40S) subunit, essential for control of translational accuracy; has similarity to E. coli S5 and rat S2 ribosomal proteins |
|
| YLR048W | 36.51 |
RPS0B
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Ap; required for maturation of 18S rRNA along with Rps0Ap; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YGR108W | 36.40 |
CLB1
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YNL162W | 35.64 |
RPL42A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Bp and has similarity to rat L44 ribosomal protein |
|
| YER074W | 35.33 |
RPS24A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Bp and has similarity to rat S24 ribosomal protein |
|
| YLR029C | 35.24 |
RPL15A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Bp and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YBR191W | 35.01 |
RPL21A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl21Bp and has similarity to rat L21 ribosomal protein |
|
| YDL136W | 34.91 |
RPL35B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl35Ap and has similarity to rat L35 ribosomal protein |
|
| YKR092C | 34.20 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YLL045C | 34.11 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YDL191W | 34.10 |
RPL35A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl35Bp and has similarity to rat L35 ribosomal protein |
|
| YHR021C | 33.55 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YMR106C | 33.35 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YKL180W | 33.32 |
RPL17A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl17Bp and has similarity to E. coli L22 and rat L17 ribosomal proteins; copurifies with the Dam1 complex (aka DASH complex) |
|
| YNL339C | 32.56 |
YRF1-6
|
Helicase encoded by the Y' element of subtelomeric regions, highly expressed in the mutants lacking the telomerase component TLC1; potentially phosphorylated by Cdc28p |
|
| YDR450W | 32.35 |
RPS18A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Bp and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YIL053W | 32.25 |
RHR2
|
Constitutively expressed isoform of DL-glycerol-3-phosphatase; involved in glycerol biosynthesis, induced in response to both anaerobic and, along with the Hor2p/Gpp2p isoform, osmotic stress |
|
| YHR203C | 32.20 |
RPS4B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps4Ap and has similarity to rat S4 ribosomal protein |
|
| YLR325C | 32.10 |
RPL38
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L38 ribosomal protein |
|
| YOL120C | 31.51 |
RPL18A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl18Bp and has similarity to rat L18 ribosomal protein; intron of RPL18A pre-mRNA forms stem-loop structures that are a target for Rnt1p cleavage leading to degradation |
|
| YDL075W | 31.46 |
RPL31A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Bp and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YMR116C | 31.45 |
ASC1
|
G-protein beta subunit and guanine nucleotide dissociation inhibitor for Gpa2p; ortholog of RACK1 that inhibits translation; core component of the small (40S) ribosomal subunit; represses Gcn4p in the absence of amino acid starvation |
|
| YOR101W | 30.88 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YGR085C | 30.58 |
RPL11B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl11Ap; involved in ribosomal assembly; depletion causes degradation of proteins and RNA of the 60S subunit; has similarity to E. coli L5 and rat L11 |
|
| YNL178W | 30.24 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YFL034C-A | 30.22 |
RPL22B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl22Ap and to rat L22 ribosomal protein |
|
| YNR013C | 29.56 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YGL031C | 29.55 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YJR123W | 29.24 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YGL147C | 29.06 |
RPL9A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl9Bp and has similarity to E. coli L6 and rat L9 ribosomal proteins |
|
| YLR154W-B | 29.06 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YHR010W | 29.04 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YBL027W | 28.84 |
RPL19B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Ap and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YLR154W-A | 28.76 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR110C | 28.32 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YLR344W | 28.23 |
RPL26A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Bp and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YKL030W | 28.17 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene MAE1 |
||
| YDL055C | 27.88 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YER031C | 27.84 |
YPT31
|
GTPase of the Ypt/Rab family, very similar to Ypt32p; involved in the exocytic pathway; mediates intra-Golgi traffic or the budding of post-Golgi vesicles from the trans-Golgi |
|
| YNL336W | 27.58 |
COS1
|
Protein of unknown function, member of the DUP380 subfamily of conserved, often subtelomerically-encoded proteins |
|
| YBR085W | 27.26 |
AAC3
|
Mitochondrial inner membrane ADP/ATP translocator, exchanges cytosolic ADP for mitochondrially synthesized ATP; expressed under anaerobic conditions; similar to Pet9p and Aac1p; has roles in maintenance of viability and in respiration |
|
| YPL131W | 26.93 |
RPL5
|
Protein component of the large (60S) ribosomal subunit with similarity to E. coli L18 and rat L5 ribosomal proteins; binds 5S rRNA and is required for 60S subunit assembly |
|
| YLR400W | 26.83 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFR032C-A | 26.52 |
RPL29
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L29 ribosomal protein; not essential for translation, but required for proper joining of the large and small ribosomal subunits and for normal translation rate |
|
| YOR312C | 26.31 |
RPL20B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl20Ap and has similarity to rat L18a ribosomal protein |
|
| YGR052W | 25.67 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YMR194W | 25.57 |
RPL36A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl36Bp and has similarity to rat L36 ribosomal protein; binds to 5.8 S rRNA |
|
| YPL143W | 25.47 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YER102W | 25.35 |
RPS8B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps8Ap and has similarity to rat S8 ribosomal protein |
|
| YBL028C | 25.34 |
Protein of unknown function that may interact with ribosomes; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus; predicted to be involved in ribosome biogenesis |
||
| YOL039W | 25.33 |
RPP2A
|
Ribosomal protein P2 alpha, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YBL087C | 25.11 |
RPL23A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl23Bp and has similarity to E. coli L14 and rat L23 ribosomal proteins |
|
| YGL030W | 25.04 |
RPL30
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L30 ribosomal protein; involved in pre-rRNA processing in the nucleolus; autoregulates splicing of its transcript |
|
| YLR333C | 24.17 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YLR312W-A | 24.02 |
MRPL15
|
Mitochondrial ribosomal protein of the large subunit |
|
| YAL038W | 23.64 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YHL033C | 23.63 |
RPL8A
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Bp and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YMR141W-A | 23.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPL13B/YMR142C |
||
| YGR259C | 23.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF TNA1/YGR260W |
||
| YOL040C | 22.94 |
RPS15
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S19 and rat S15 ribosomal proteins |
|
| YNL096C | 22.67 |
RPS7B
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Ap; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YGR050C | 22.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHL001W | 22.56 |
RPL14B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl14Ap and has similarity to rat L14 ribosomal protein |
|
| YFL015W-A | 22.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR339C | 22.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the essential gene RPP0 |
||
| YDR279W | 22.04 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YAL003W | 21.87 |
EFB1
|
Translation elongation factor 1 beta; stimulates nucleotide exchange to regenerate EF-1 alpha-GTP for the next elongation cycle; part of the EF-1 complex, which facilitates binding of aminoacyl-tRNA to the ribosomal A site |
|
| YCR031C | 21.39 |
RPS14A
|
Ribosomal protein 59 of the small subunit, required for ribosome assembly and 20S pre-rRNA processing; mutations confer cryptopleurine resistance; nearly identical to Rps14Bp and similar to E. coli S11 and rat S14 ribosomal proteins |
|
| YGR060W | 21.32 |
ERG25
|
C-4 methyl sterol oxidase, catalyzes the first of three steps required to remove two C-4 methyl groups from an intermediate in ergosterol biosynthesis; mutants accumulate the sterol intermediate 4,4-dimethylzymosterol |
|
| YLR340W | 21.09 |
RPP0
|
Conserved ribosomal protein P0 similar to rat P0, human P0, and E. coli L10e; shown to be phosphorylated on serine 302 |
|
| YIL148W | 20.98 |
RPL40A
|
Fusion protein, identical to Rpl40Bp, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YFL015C | 20.92 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YFL022C | 20.89 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YLR075W | 20.79 |
RPL10
|
Protein component of the large (60S) ribosomal subunit, responsible for joining the 40S and 60S subunits; regulates translation initiation; has similarity to rat L10 ribosomal protein and to members of the QM gene family |
|
| YHR181W | 20.78 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YMR230W | 20.59 |
RPS10B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps10Ap and has similarity to rat ribosomal protein S10 |
|
| YDR500C | 19.94 |
RPL37B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl37Ap and to rat L37 ribosomal protein |
|
| YLR154W-C | 19.88 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YMR083W | 19.56 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YKR057W | 19.43 |
RPS21A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Bp and has similarity to rat S21 ribosomal protein |
|
| YLR185W | 19.39 |
RPL37A
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl37Bp and to rat L37 ribosomal protein |
|
| YPL249C-A | 19.28 |
RPL36B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl36Ap and has similarity to rat L36 ribosomal protein; binds to 5.8 S rRNA |
|
| YKL152C | 19.05 |
GPM1
|
Tetrameric phosphoglycerate mutase, mediates the conversion of 3-phosphoglycerate to 2-phosphoglycerate during glycolysis and the reverse reaction during gluconeogenesis |
|
| YLR183C | 18.95 |
TOS4
|
Transcription factor that binds to a number of promoter regions, particularly promoters of some genes involved in pheromone response and cell cycle; potential Cdc28p substrate; expression is induced in G1 by bound SBF |
|
| YEL068C | 18.95 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL085C | 18.72 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YOR369C | 18.22 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YFR054C | 17.57 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER032W | 17.56 |
FIR1
|
Protein involved in 3' mRNA processing, interacts with Ref2p; potential Cdc28p substrate |
|
| YGR192C | 17.50 |
TDH3
|
Glyceraldehyde-3-phosphate dehydrogenase, isozyme 3, involved in glycolysis and gluconeogenesis; tetramer that catalyzes the reaction of glyceraldehyde-3-phosphate to 1,3 bis-phosphoglycerate; detected in the cytoplasm and cell-wall |
|
| YMR183C | 17.43 |
SSO2
|
Plasma membrane t-SNARE involved in fusion of secretory vesicles at the plasma membrane; syntaxin homolog that is functionally redundant with Sso1p |
|
| YMR205C | 17.41 |
PFK2
|
Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes |
|
| YER130C | 17.14 |
Hypothetical protein |
||
| YOR315W | 17.05 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YJR094C | 16.98 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YGR051C | 16.92 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YDR041W | 16.86 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YKL081W | 16.60 |
TEF4
|
Translation elongation factor EF-1 gamma |
|
| YHR142W | 16.57 |
CHS7
|
Protein of unknown function, involved in chitin biosynthesis by regulating Chs3p export from the ER |
|
| YGR234W | 16.56 |
YHB1
|
Nitric oxide oxidoreductase, flavohemoglobin involved in nitric oxide detoxification; plays a role in the oxidative and nitrosative stress responses |
|
| YLR003C | 16.46 |
Putative protein of unknown function that may participate in DNA replication; green fluorescent protein (GFP)-fusion protein is localized to the nucleus; YLR003C is not an essential gene |
||
| YHR032W-A | 16.44 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YHR032C-A |
||
| YGR253C | 16.27 |
PUP2
|
Alpha 5 subunit of the 20S proteasome involved in ubiquitin-dependent catabolism; human homolog is subunit zeta |
|
| YDR188W | 16.20 |
CCT6
|
Subunit of the cytosolic chaperonin Cct ring complex, related to Tcp1p, essential protein that is required for the assembly of actin and tubulins in vivo; contains an ATP-binding motif |
|
| YPR063C | 16.10 |
ER-localized protein of unknown function |
||
| YDR449C | 16.08 |
UTP6
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YCL024W | 15.99 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YNL024C | 15.98 |
Putative protein of unknown function with seven beta-strand methyltransferase motif; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YNL024C is not an essential gene |
||
| YDR037W | 15.88 |
KRS1
|
Lysyl-tRNA synthetase; also identified as a negative regulator of general control of amino acid biosynthesis |
|
| YDR133C | 15.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR134C |
||
| YLR388W | 15.78 |
RPS29A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps29Bp and has similarity to rat S29 and E. coli S14 ribosomal proteins |
|
| YLR300W | 15.65 |
EXG1
|
Major exo-1,3-beta-glucanase of the cell wall, involved in cell wall beta-glucan assembly; exists as three differentially glycosylated isoenzymes |
|
| YLR287C-A | 15.48 |
RPS30A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps30Bp and has similarity to rat S30 ribosomal protein |
|
| YDL023C | 15.35 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YEL056W | 15.24 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YLR401C | 15.18 |
DUS3
|
Dihydrouridine synthase, member of a widespread family of conserved proteins including Smm1p, Dus1p, and Dus4p; contains a consensus oleate response element (ORE) in its promoter region |
|
| YGR260W | 15.12 |
TNA1
|
High affinity nicotinic acid plasma membrane permease, responsible for uptake of low levels of nicotinic acid; expression of the gene increases in the absence of extracellular nicotinic acid or para-aminobenzoate (PABA) |
|
| YDR044W | 15.09 |
HEM13
|
Coproporphyrinogen III oxidase, an oxygen requiring enzyme that catalyzes the sixth step in the heme biosynthetic pathway; localizes to the mitochondrial inner membrane; transcription is repressed by oxygen and heme (via Rox1p and Hap1p) |
|
| YPR080W | 14.99 |
TEF1
|
Translational elongation factor EF-1 alpha; also encoded by TEF2; functions in the binding reaction of aminoacyl-tRNA (AA-tRNA) to ribosomes |
|
| YPL227C | 14.98 |
ALG5
|
UDP-glucose:dolichyl-phosphate glucosyltransferase, involved in asparagine-linked glycosylation in the endoplasmic reticulum |
|
| YPR029C | 14.96 |
APL4
|
Gamma-adaptin, large subunit of the clathrin-associated protein (AP-1) complex; binds clathrin; involved in vesicle mediated transport |
|
| YHR180W-A | 14.95 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YDR040C | 14.74 |
ENA1
|
P-type ATPase sodium pump, involved in Na+ and Li+ efflux to allow salt tolerance |
|
| YAL033W | 14.73 |
POP5
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YBR190W | 14.71 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the verified ribosomal protein gene RPL21A/YBR191W |
||
| YNL302C | 14.71 |
RPS19B
|
Protein component of the small (40S) ribosomal subunit, required for assembly and maturation of pre-40 S particles; mutations in human RPS19 are associated with Diamond Blackfan anemia; nearly identical to Rps19Ap |
|
| YDR187C | 14.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified, essential ORF CCT6/YDR188W |
||
| YDR112W | 14.61 |
IRC2
|
Dubious open reading frame, unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR111C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YML064C | 14.57 |
TEM1
|
GTP-binding protein of the ras superfamily involved in termination of M-phase; controls actomyosin and septin dynamics during cytokinesis |
|
| YFR028C | 14.39 |
CDC14
|
Protein phosphatase required for mitotic exit; located in the nucleolus until liberated by the FEAR and Mitotic Exit Network in anaphase, enabling it to act on key substrates to effect a decrease in CDK/B-cyclin activity and mitotic exit |
|
| YEL066W | 14.24 |
HPA3
|
D-Amino acid N-acetyltransferase, catalyzes N-acetylation of D-amino acids through ordered bi-bi mechanism in which acetyl-CoA is first substrate bound and CoA is last product liberated; similar to Hpa2p, acetylates histones weakly in vitro |
|
| YLR249W | 14.18 |
YEF3
|
Translational elongation factor 3, stimulates the binding of aminoacyl-tRNA (AA-tRNA) to ribosomes by releasing EF-1 alpha from the ribosomal complex; contains two ABC cassettes; binds and hydrolyses ATP |
|
| YPR131C | 14.12 |
NAT3
|
Catalytic subunit of the NatB N-terminal acetyltransferase, which catalyzes acetylation of the amino-terminal methionine residues of all proteins beginning with Met-Asp or Met-Glu and of some proteins beginning with Met-Asn or Met-Met |
Network of associatons between targets according to the STRING database.
First level regulatory network of RAP1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 39.7 | 158.6 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 36.8 | 147.1 | GO:0046898 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 27.1 | 434.4 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 25.5 | 3497.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 23.1 | 69.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 13.3 | 40.0 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 10.7 | 32.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 10.2 | 143.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 10.2 | 30.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 9.3 | 27.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 8.5 | 59.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 8.3 | 83.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 7.6 | 30.6 | GO:0071331 | cellular glucose homeostasis(GO:0001678) carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 7.2 | 35.8 | GO:0007535 | donor selection(GO:0007535) |
| 7.1 | 28.3 | GO:0098740 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 7.0 | 20.9 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 6.8 | 27.3 | GO:0015886 | heme transport(GO:0015886) |
| 5.7 | 17.1 | GO:0046039 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 5.7 | 51.0 | GO:0006814 | sodium ion transport(GO:0006814) |
| 5.1 | 35.9 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 4.7 | 14.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 4.7 | 113.7 | GO:0006414 | translational elongation(GO:0006414) |
| 4.7 | 18.9 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 4.6 | 13.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) peptidyl-methionine modification(GO:0018206) |
| 4.5 | 13.4 | GO:0038032 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 4.0 | 15.9 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 3.9 | 19.3 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 3.9 | 27.0 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 3.9 | 11.6 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 3.8 | 26.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 3.6 | 102.1 | GO:0006096 | glycolytic process(GO:0006096) nucleoside diphosphate phosphorylation(GO:0006165) ATP generation from ADP(GO:0006757) |
| 3.5 | 14.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 3.3 | 13.4 | GO:0015909 | long-chain fatty acid transport(GO:0015909) fatty-acyl-CoA metabolic process(GO:0035337) |
| 3.3 | 16.4 | GO:0046417 | chorismate metabolic process(GO:0046417) |
| 3.3 | 9.8 | GO:1901070 | GMP biosynthetic process(GO:0006177) guanosine-containing compound biosynthetic process(GO:1901070) |
| 3.2 | 9.7 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 3.1 | 9.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 3.1 | 15.4 | GO:0035392 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 3.0 | 21.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 3.0 | 9.0 | GO:0035952 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 2.9 | 5.9 | GO:0046083 | adenine metabolic process(GO:0046083) |
| 2.9 | 28.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 2.8 | 13.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 2.8 | 8.3 | GO:1902101 | positive regulation of mitotic nuclear division(GO:0045840) positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 2.7 | 2.7 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 2.7 | 10.8 | GO:0090338 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 2.7 | 48.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 2.7 | 10.6 | GO:0042938 | dipeptide transport(GO:0042938) |
| 2.6 | 13.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 2.6 | 7.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 2.5 | 2.5 | GO:0010695 | regulation of spindle pole body separation(GO:0010695) |
| 2.5 | 9.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 2.4 | 7.3 | GO:0071478 | cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 2.4 | 21.8 | GO:0071966 | cell wall chitin biosynthetic process(GO:0006038) fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 2.4 | 7.1 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 2.3 | 11.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 2.2 | 11.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 2.2 | 30.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 2.0 | 10.1 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 1.9 | 9.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.9 | 21.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 1.9 | 11.4 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 1.9 | 9.4 | GO:0006032 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 1.8 | 12.8 | GO:0000376 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 1.7 | 13.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.6 | 8.2 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 1.6 | 6.5 | GO:0000296 | spermine transport(GO:0000296) |
| 1.6 | 11.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 1.6 | 42.4 | GO:0044108 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 1.5 | 20.9 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 1.5 | 1.5 | GO:0070592 | cell wall polysaccharide biosynthetic process(GO:0070592) |
| 1.5 | 51.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 1.5 | 5.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 1.4 | 7.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 1.4 | 12.9 | GO:0031070 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 1.4 | 1.4 | GO:0006901 | vesicle coating(GO:0006901) |
| 1.4 | 42.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 1.4 | 5.7 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 1.4 | 19.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 1.4 | 7.0 | GO:0009371 | positive regulation of transcription from RNA polymerase II promoter by pheromones(GO:0007329) positive regulation of transcription by pheromones(GO:0009371) |
| 1.4 | 4.1 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 1.4 | 4.1 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) regulation of cellular amine catabolic process(GO:0033241) |
| 1.3 | 5.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 1.3 | 4.0 | GO:0051238 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 1.3 | 5.2 | GO:0006563 | L-serine metabolic process(GO:0006563) |
| 1.3 | 9.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.3 | 28.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 1.3 | 10.2 | GO:0090522 | vesicle tethering involved in exocytosis(GO:0090522) |
| 1.2 | 2.5 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 1.2 | 7.3 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 1.2 | 4.7 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 1.2 | 10.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 1.2 | 15.1 | GO:0000147 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 1.1 | 20.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 1.1 | 5.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 1.1 | 3.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 1.1 | 13.9 | GO:0006862 | nucleotide transport(GO:0006862) cofactor transport(GO:0051181) |
| 1.1 | 6.4 | GO:0070941 | eisosome assembly(GO:0070941) |
| 1.1 | 3.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 1.0 | 5.2 | GO:0007119 | budding cell isotropic bud growth(GO:0007119) |
| 1.0 | 11.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 1.0 | 3.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.0 | 3.0 | GO:1902707 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 1.0 | 14.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 1.0 | 11.9 | GO:0009636 | response to toxic substance(GO:0009636) |
| 1.0 | 3.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 1.0 | 7.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 1.0 | 23.8 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.9 | 2.7 | GO:0044109 | cellular alcohol catabolic process(GO:0044109) positive regulation of transcription from RNA polymerase II promoter in response to amino acid starvation(GO:0061412) |
| 0.9 | 8.7 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.9 | 4.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.8 | 3.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.8 | 7.3 | GO:0051320 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.8 | 0.8 | GO:0031555 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) transcriptional attenuation(GO:0031555) transcription antitermination(GO:0031564) |
| 0.8 | 3.2 | GO:0006560 | proline metabolic process(GO:0006560) proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.8 | 11.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.8 | 0.8 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.7 | 18.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.7 | 2.2 | GO:0042992 | negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.7 | 5.7 | GO:0006903 | vesicle targeting(GO:0006903) vesicle localization(GO:0051648) establishment of vesicle localization(GO:0051650) |
| 0.7 | 11.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.7 | 4.3 | GO:0030847 | termination of RNA polymerase II transcription, exosome-dependent(GO:0030847) |
| 0.7 | 4.2 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.7 | 4.9 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.7 | 4.6 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.6 | 4.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.6 | 70.9 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.6 | 2.3 | GO:0019365 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.6 | 1.1 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.6 | 1.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.6 | 3.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.5 | 1.6 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.5 | 1.6 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.5 | 8.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.5 | 8.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.5 | 3.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.5 | 9.3 | GO:0051647 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.5 | 16.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.5 | 11.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.5 | 10.0 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.4 | 2.7 | GO:0009097 | isoleucine biosynthetic process(GO:0009097) |
| 0.4 | 1.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.4 | 14.1 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.4 | 4.8 | GO:0043094 | cellular metabolic compound salvage(GO:0043094) |
| 0.4 | 3.5 | GO:0006189 | IMP biosynthetic process(GO:0006188) 'de novo' IMP biosynthetic process(GO:0006189) IMP metabolic process(GO:0046040) |
| 0.4 | 2.2 | GO:0048309 | endoplasmic reticulum inheritance(GO:0048309) |
| 0.4 | 7.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.4 | 0.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.4 | 1.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.4 | 1.2 | GO:0000730 | meiotic DNA recombinase assembly(GO:0000707) DNA recombinase assembly(GO:0000730) |
| 0.4 | 11.3 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.4 | 5.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.4 | 1.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 1.9 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.4 | 3.0 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.4 | 0.7 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.4 | 10.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.4 | 2.1 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.3 | 4.3 | GO:0007117 | budding cell bud growth(GO:0007117) |
| 0.3 | 1.0 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.3 | 6.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.3 | 2.5 | GO:0000921 | septin ring assembly(GO:0000921) |
| 0.3 | 2.7 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 1.6 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.3 | 6.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.3 | 1.1 | GO:0046655 | folic acid metabolic process(GO:0046655) folic acid biosynthetic process(GO:0046656) |
| 0.3 | 3.6 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.2 | 1.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 15.0 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.2 | 4.6 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.2 | 1.8 | GO:0010529 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.2 | 0.5 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) barbed-end actin filament capping(GO:0051016) actin filament capping(GO:0051693) |
| 0.2 | 1.5 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.2 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 10.4 | GO:0051302 | regulation of cell division(GO:0051302) |
| 0.2 | 0.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 0.9 | GO:0070058 | tRNA gene clustering(GO:0070058) |
| 0.2 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.6 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.1 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 2.5 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 1.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.5 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.0 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.1 | 0.9 | GO:0031120 | tRNA pseudouridine synthesis(GO:0031119) snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.1 | 0.6 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.1 | 1.0 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 2.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.5 | GO:0046219 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.1 | 0.2 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.1 | 2.4 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.1 | 0.5 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 | 0.3 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 0.4 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.1 | 0.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.1 | 0.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.4 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.1 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.0 | 1.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.4 | GO:0010833 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.2 | 1902.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 29.5 | 2480.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 19.9 | 59.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 18.4 | 92.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 13.3 | 40.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 6.2 | 18.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 4.8 | 14.4 | GO:0030869 | RENT complex(GO:0030869) |
| 4.2 | 59.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 4.2 | 33.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 3.9 | 11.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 3.0 | 18.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 2.9 | 34.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 2.9 | 11.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.7 | 16.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 2.7 | 10.7 | GO:0033179 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 2.6 | 7.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 2.5 | 15.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 2.5 | 7.5 | GO:0071261 | Ssh1 translocon complex(GO:0071261) |
| 2.3 | 16.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.2 | 6.7 | GO:0097344 | Rix1 complex(GO:0097344) |
| 2.2 | 15.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 2.1 | 8.3 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 2.0 | 13.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 1.9 | 9.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.9 | 32.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 1.9 | 5.6 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.8 | 14.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 1.8 | 5.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 1.8 | 5.3 | GO:0030428 | cell septum(GO:0030428) |
| 1.7 | 13.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 1.7 | 3.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.7 | 15.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 1.5 | 12.2 | GO:0042597 | periplasmic space(GO:0042597) |
| 1.5 | 11.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.5 | 5.9 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 1.4 | 12.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 1.4 | 13.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 1.3 | 22.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 1.3 | 14.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 1.3 | 10.2 | GO:0000145 | exocyst(GO:0000145) |
| 1.3 | 19.1 | GO:0005940 | septin ring(GO:0005940) |
| 1.2 | 22.5 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 1.2 | 35.7 | GO:0005844 | polysome(GO:0005844) |
| 1.2 | 53.6 | GO:0044445 | cytosolic part(GO:0044445) |
| 1.2 | 4.7 | GO:0000938 | GARP complex(GO:0000938) |
| 1.2 | 4.6 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 1.1 | 8.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.1 | 3.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.0 | 7.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 1.0 | 4.8 | GO:0031588 | nuclear envelope lumen(GO:0005641) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.9 | 4.6 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.9 | 2.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.9 | 42.8 | GO:0030659 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) |
| 0.9 | 4.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.9 | 5.2 | GO:0032126 | eisosome(GO:0032126) |
| 0.8 | 4.0 | GO:0000500 | RNA polymerase I upstream activating factor complex(GO:0000500) |
| 0.7 | 5.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.7 | 15.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.7 | 10.0 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.7 | 4.0 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.7 | 23.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.7 | 26.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 11.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.6 | 5.1 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.6 | 2.5 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.6 | 96.9 | GO:0005933 | cellular bud(GO:0005933) |
| 0.6 | 4.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.6 | 18.8 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.6 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.6 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.6 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 3.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 35.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.5 | 1.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.5 | 3.0 | GO:0070772 | PAS complex(GO:0070772) |
| 0.5 | 7.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.4 | 18.9 | GO:0030479 | actin cortical patch(GO:0030479) endocytic patch(GO:0061645) |
| 0.4 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.4 | 14.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.4 | 3.6 | GO:0098589 | membrane region(GO:0098589) |
| 0.4 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.3 | 1.0 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 15.4 | GO:0005815 | microtubule organizing center(GO:0005815) spindle pole body(GO:0005816) |
| 0.3 | 4.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 1.3 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.3 | 1.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 1.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 2.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 2.7 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.2 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.8 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 22.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.2 | 0.9 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.2 | 1.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 0.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.2 | 1.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 0.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 0.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 1.6 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.2 | 10.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.6 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 1.1 | GO:0034708 | methyltransferase complex(GO:0034708) |
| 0.1 | 6.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 4.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.8 | 4354.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 19.9 | 59.8 | GO:0019003 | GDP binding(GO:0019003) |
| 14.9 | 59.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 10.1 | 60.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 9.9 | 29.7 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 9.7 | 29.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 9.0 | 26.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 8.1 | 48.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 7.4 | 37.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 7.0 | 21.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 7.0 | 20.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 6.8 | 27.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 5.2 | 15.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 5.0 | 24.9 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 4.5 | 17.8 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 4.5 | 57.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 4.4 | 44.0 | GO:0005216 | ion channel activity(GO:0005216) |
| 4.3 | 21.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 4.3 | 21.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 4.2 | 59.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 4.1 | 16.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 4.0 | 15.9 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 3.9 | 11.8 | GO:0004619 | phosphoglycerate mutase activity(GO:0004619) |
| 3.9 | 11.6 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 3.9 | 15.5 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 3.9 | 11.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 3.8 | 11.4 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 3.8 | 15.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 3.5 | 17.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 3.5 | 13.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 3.3 | 13.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 3.3 | 16.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 3.1 | 18.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 2.9 | 17.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 2.9 | 11.6 | GO:0016725 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.7 | 31.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 2.6 | 13.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 2.5 | 15.3 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 2.5 | 7.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 2.3 | 9.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 2.0 | 6.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.9 | 65.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 1.9 | 5.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.9 | 5.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 1.9 | 11.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 1.9 | 7.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.8 | 5.3 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 1.8 | 3.5 | GO:0099516 | ion antiporter activity(GO:0099516) |
| 1.8 | 3.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.7 | 16.9 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 1.7 | 16.9 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 1.7 | 6.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 1.6 | 6.5 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 1.6 | 6.4 | GO:1901677 | phosphate transmembrane transporter activity(GO:1901677) |
| 1.6 | 12.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 1.6 | 32.8 | GO:0019901 | protein kinase binding(GO:0019901) |
| 1.5 | 4.6 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 1.5 | 6.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 1.5 | 13.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 1.5 | 16.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 1.5 | 11.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.4 | 12.5 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 1.4 | 13.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 1.3 | 9.4 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 1.3 | 15.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 1.3 | 12.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) ribonuclease P activity(GO:0004526) |
| 1.3 | 7.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 1.3 | 15.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 1.3 | 25.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 1.2 | 25.3 | GO:0051287 | NAD binding(GO:0051287) |
| 1.2 | 22.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 1.2 | 73.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 1.0 | 3.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 1.0 | 4.0 | GO:0001168 | transcription factor activity, RNA polymerase I upstream control element sequence-specific binding(GO:0001168) |
| 1.0 | 4.0 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 1.0 | 8.7 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.9 | 2.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.9 | 9.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.9 | 20.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.8 | 2.5 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.8 | 5.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.8 | 5.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.8 | 6.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.8 | 24.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.8 | 11.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.8 | 5.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.8 | 3.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.7 | 2.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.7 | 13.2 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.7 | 18.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.7 | 10.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.7 | 5.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.7 | 8.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.7 | 2.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.7 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.7 | 3.4 | GO:0042124 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.7 | 48.0 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.6 | 4.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.6 | 41.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.6 | 5.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.5 | 10.4 | GO:0016769 | transaminase activity(GO:0008483) transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 1.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.5 | 10.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 2.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.5 | 1.9 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.5 | 19.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.4 | 2.2 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.4 | 1.6 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.4 | 2.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.4 | 4.6 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.4 | 5.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.4 | 2.3 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.4 | 13.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.4 | 2.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 1.7 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 9.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.3 | 10.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 1.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.8 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.3 | 3.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 14.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.3 | 4.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.3 | 6.0 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.3 | 2.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.2 | 1.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.2 | 5.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 2.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 2.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.2 | 1.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 6.4 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.2 | 55.1 | GO:0003723 | RNA binding(GO:0003723) |
| 0.2 | 0.8 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 1.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 3.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.2 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.2 | 0.6 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.1 | 1.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.4 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 1.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 1.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 0.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.7 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.1 | 3.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 2.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.3 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) |
| 0.1 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.2 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0032934 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0035591 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 20.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 3.5 | 10.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 1.7 | 8.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 1.4 | 5.4 | PID E2F PATHWAY | E2F transcription factor network |
| 1.3 | 3.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.8 | 2.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.6 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.8 | 254.1 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 31.0 | 496.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 7.0 | 20.9 | REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | Genes involved in Insulin receptor signalling cascade |
| 5.5 | 16.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 5.4 | 21.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 4.3 | 12.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 4.2 | 8.3 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 3.1 | 12.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 3.0 | 9.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 2.6 | 7.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 1.9 | 15.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.3 | 1.3 | REACTOME METABOLISM OF MRNA | Genes involved in Metabolism of mRNA |
| 0.8 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.6 | 2.3 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.5 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.5 | 3.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.4 | 4.6 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.3 | 1.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.2 | 1.6 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.2 | 2.6 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.1 | 0.3 | REACTOME DEVELOPMENTAL BIOLOGY | Genes involved in Developmental Biology |