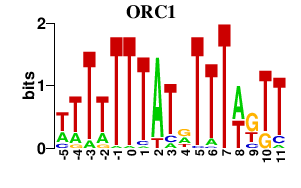
Results for ORC1
Z-value: 1.04
Transcription factors associated with ORC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ORC1
|
S000004530 | Largest subunit of the origin recognition complex |
Activity-expression correlation:
Activity profile of ORC1 motif
Sorted Z-values of ORC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YAR053W | 9.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR244W | 5.58 |
Putative protein of unknown function |
||
| YAR060C | 5.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR311C | 5.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR212C | 4.66 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR001W | 4.41 |
CIT3
|
Dual specificity mitochondrial citrate and methylcitrate synthase; catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate and that of propionyl-CoA and oxaloacetate to form 2-methylcitrate |
|
| YHR212W-A | 4.14 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YER065C | 3.86 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YBR200W-A | 3.86 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YAL039C | 3.72 |
CYC3
|
Cytochrome c heme lyase (holocytochrome c synthase), attaches heme to apo-cytochrome c (Cyc1p or Cyc7p) in the mitochondrial intermembrane space; human ortholog may have a role in microphthalmia with linear skin defects (MLS) |
|
| YMR107W | 3.68 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YAL062W | 3.57 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YNL013C | 3.52 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF HEF3/YNL014W |
||
| YOR345C | 3.43 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YAR070C | 3.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YCL054W | 3.31 |
SPB1
|
AdoMet-dependent methyltransferase involved in rRNA processing and 60S ribosomal subunit maturation; methylates G2922 in the tRNA docking site of the large subunit rRNA and in the absence of snR52, U2921; suppressor of PAB1 mutants |
|
| YAR069C | 3.12 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR138C | 3.11 |
Putative protein of unknown function; has similarity to Pbi2p; double null mutant lacking Pbi2p and Yhr138p exhibits highly fragmented vacuoles |
||
| YKL217W | 3.03 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YOR346W | 2.99 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YNL180C | 2.83 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YAR050W | 2.81 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YCL069W | 2.79 |
VBA3
|
Permease of basic amino acids in the vacuolar membrane |
|
| YFR032C-B | 2.79 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YGR225W | 2.71 |
AMA1
|
Activator of meiotic anaphase promoting complex (APC/C); Cdc20p family member; required for initiation of spore wall assembly; required for Clb1p degradation during meiosis |
|
| YJR095W | 2.69 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YNL179C | 2.65 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YJR048W | 2.62 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YIL057C | 2.59 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YKR105C | 2.51 |
VBA5
|
Putative transporter of the Major Facilitator Superfamily (MFS); proposed role as a basic amino acid permease based on phylogeny |
|
| YBR296C | 2.49 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YPL033C | 2.49 |
Putative protein of unknown function; may be involved in DNA metabolism; expression is induced by Kar4p |
||
| YGR087C | 2.43 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YEL030C-A | 2.42 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YDL210W | 2.41 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YML042W | 2.37 |
CAT2
|
Carnitine acetyl-CoA transferase present in both mitochondria and peroxisomes, transfers activated acetyl groups to carnitine to form acetylcarnitine which can be shuttled across membranes |
|
| YIL055C | 2.37 |
Putative protein of unknown function |
||
| YEL030W | 2.29 |
ECM10
|
Heat shock protein of the Hsp70 family, localized in mitochondrial nucleoids, plays a role in protein translocation, interacts with Mge1p in an ATP-dependent manner; overexpression induces extensive mitochondrial DNA aggregations |
|
| YDL244W | 2.29 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YOL051W | 2.28 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YLR307W | 2.26 |
CDA1
|
Chitin deacetylase, together with Cda2p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YBL043W | 2.25 |
ECM13
|
Non-essential protein of unknown function; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YMR311C | 2.25 |
GLC8
|
Regulatory subunit of protein phosphatase 1 (Glc7p), involved in glycogen metabolism and chromosome segregation; proposed to regulate Glc7p activity via conformational alteration; ortholog of the mammalian protein phosphatase inhibitor 2 |
|
| YMR206W | 2.23 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YKR097W | 2.21 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YGR236C | 2.13 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR307C-A | 2.13 |
Putative protein of unknown function |
||
| YDL138W | 2.13 |
RGT2
|
Plasma membrane glucose receptor, highly similar to Snf3p; both Rgt2p and Snf3p serve as transmembrane glucose sensors generating an intracellular signal that induces expression of glucose transporter (HXT) genes |
|
| YDR259C | 2.11 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YGR258C | 2.09 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YDL139C | 2.04 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YFL011W | 2.04 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YBR292C | 1.99 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YDL223C | 1.98 |
HBT1
|
Substrate of the Hub1p ubiquitin-like protein that localizes to the shmoo tip (mating projection); mutants are defective for mating projection formation, thereby implicating Hbt1p in polarized cell morphogenesis |
|
| YGL163C | 1.98 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YLR356W | 1.97 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YKL031W | 1.97 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species |
||
| YLR296W | 1.93 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YEL007W | 1.92 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YOR214C | 1.92 |
Putative protein of unknown function; YOR214C is not an essential gene |
||
| YAR047C | 1.91 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER033C | 1.89 |
ZRG8
|
Protein of unknown function; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; GFP-fusion protein is localized to the cytoplasm; transcription induced under conditions of zinc deficiency |
|
| YKL109W | 1.83 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YFL051C | 1.82 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YAL054C | 1.80 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YLR327C | 1.80 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YJL037W | 1.78 |
IRC18
|
Putative protein of unknown function; expression induced in respiratory-deficient cells and in carbon-limited chemostat cultures; similar to adjacent ORF, YJL038C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YOL060C | 1.77 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YNL093W | 1.73 |
YPT53
|
GTPase, similar to Ypt51p and Ypt52p and to mammalian rab5; required for vacuolar protein sorting and endocytosis |
|
| YGR144W | 1.72 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YDR342C | 1.67 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YHR139C | 1.67 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YGL096W | 1.66 |
TOS8
|
Homeodomain-containing transcription factor; SBF regulated target gene that in turn regulates expression of genes involved in G1/S phase events such as bud site selection, bud emergence and cell cycle progression; similarity to Cup9p |
|
| YOR178C | 1.62 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YJR078W | 1.62 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YHR217C | 1.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YKL028W | 1.60 |
TFA1
|
TFIIE large subunit, involved in recruitment of RNA polymerase II to the promoter, activation of TFIIH, and promoter opening |
|
| YCL038C | 1.58 |
ATG22
|
Vacuolar integral membrane protein required for efflux of amino acids during autophagic body breakdown in the vacuole; null mutation causes a gradual loss of viability during starvation |
|
| YNR073C | 1.58 |
Putative mannitol dehydrogenase |
||
| YGR059W | 1.57 |
SPR3
|
Sporulation-specific homolog of the yeast CDC3/10/11/12 family of bud neck microfilament genes; septin protein involved in sporulation; regulated by ABFI |
|
| YNL117W | 1.55 |
MLS1
|
Malate synthase, enzyme of the glyoxylate cycle, involved in utilization of non-fermentable carbon sources; expression is subject to carbon catabolite repression; localizes in peroxisomes during growth in oleic acid medium |
|
| YMR182C | 1.54 |
RGM1
|
Putative transcriptional repressor with proline-rich zinc fingers; overproduction impairs cell growth |
|
| YHR095W | 1.54 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL066C | 1.53 |
RNR3
|
One of two large regulatory subunits of ribonucleotide-diphosphate reductase; the RNR complex catalyzes rate-limiting step in dNTP synthesis, regulated by DNA replication and DNA damage checkpoint pathways via localization of small subunits |
|
| YEL012W | 1.53 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YDL204W | 1.52 |
RTN2
|
Protein of unknown function; has similarity to mammalian reticulon proteins; member of the RTNLA (reticulon-like A) subfamily |
|
| YBR250W | 1.52 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YCR091W | 1.51 |
KIN82
|
Putative serine/threonine protein kinase, most similar to cyclic nucleotide-dependent protein kinase subfamily and the protein kinase C subfamily |
|
| YJR128W | 1.51 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YER121W | 1.47 |
Putative protein of unknown function; may be involved in phosphatase regulation and/or generation of precursor metabolites and energy |
||
| YGR043C | 1.47 |
NQM1
|
Protein of unknown function; transcription is repressed by Mot1p and induced by alpha-factor and during diauxic shift; null mutant non-quiescent cells exhibit reduced reproductive capacity |
|
| YNL014W | 1.46 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YFL058W | 1.45 |
THI5
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YMR081C | 1.43 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YPR150W | 1.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene SUE1/YPR151C |
||
| YPR002W | 1.40 |
PDH1
|
Mitochondrial protein that participates in respiration, induced by diauxic shift; homologous to E. coli PrpD, may take part in the conversion of 2-methylcitrate to 2-methylisocitrate |
|
| YAL001C | 1.37 |
TFC3
|
Largest of six subunits of the RNA polymerase III transcription initiation factor complex (TFIIIC); part of the TauB domain of TFIIIC that binds DNA at the BoxB promoter sites of tRNA and similar genes; cooperates with Tfc6p in DNA binding |
|
| YER132C | 1.36 |
PMD1
|
Protein with an N-terminal kelch-like domain, putative negative regulator of early meiotic gene expression; required, with Mds3p, for growth under alkaline conditions |
|
| YKR102W | 1.36 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YNL332W | 1.34 |
THI12
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YOR072W | 1.34 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YJL043W | 1.34 |
Putative protein of unknown function; YJL043W is a non-essential gene |
||
| YCL073C | 1.33 |
Protein of unconfirmed function; displays a topology characteristic of the Major Facilitators Superfamily of membrane proteins; coding sequence 98% identical to that of YKR106W |
||
| YAL067C | 1.30 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YPR007C | 1.29 |
REC8
|
Meiosis-specific component of sister chromatid cohesion complex; maintains cohesion between sister chromatids during meiosis I; maintains cohesion between centromeres of sister chromatids until meiosis II; homolog of S. pombe Rec8p |
|
| YGR039W | 1.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YDL245C | 1.28 |
HXT15
|
Protein of unknown function with similarity to hexose transporter family members, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YIL045W | 1.28 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YOR348C | 1.28 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YDR310C | 1.28 |
SUM1
|
Transcriptional repressor required for mitotic repression of middle sporulation-specific genes; involved in telomere maintenance, regulated by the pachytene checkpoint |
|
| YLR377C | 1.27 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YPL026C | 1.27 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YFR012W | 1.27 |
Putative protein of unknown function |
||
| YDR504C | 1.27 |
SPG3
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YKR106W | 1.27 |
Protein of unconfirmed function; displays a topology characteristic of the Major Facilitators Superfamily of membrane proteins; coding sequence 98% identical to that of YCL073C |
||
| YHR071W | 1.26 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YPL116W | 1.26 |
HOS3
|
Trichostatin A-insensitive homodimeric histone deacetylase (HDAC) with specificity in vitro for histones H3, H4, H2A, and H2B; similar to Hda1p, Rpd3p, Hos1p, and Hos2p; deletion results in increased histone acetylation at rDNA repeats |
|
| YDL085W | 1.25 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YMR017W | 1.24 |
SPO20
|
Meiosis-specific subunit of the t-SNARE complex, required for prospore membrane formation during sporulation; similar to but not functionally redundant with Sec9p; SNAP-25 homolog |
|
| YPL089C | 1.24 |
RLM1
|
MADS-box transcription factor, component of the protein kinase C-mediated MAP kinase pathway involved in the maintenance of cell integrity; phosphorylated and activated by the MAP-kinase Slt2p |
|
| YKL029C | 1.24 |
MAE1
|
Mitochondrial malic enzyme, catalyzes the oxidative decarboxylation of malate to pyruvate, which is a key intermediate in sugar metabolism and a precursor for synthesis of several amino acids |
|
| YLR278C | 1.22 |
Zinc-cluster protein; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; mutant shows moderate growth defect on caffeine; YLR278C is not an essential gene |
||
| YDL130W-A | 1.22 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YOR066W | 1.22 |
MSA1
|
Activator of G1-specific transcription factors, MBF and SBF, that regulates both the timing of G1-specific gene transcription, and cell cycle initiation; potential Cdc28p substrate |
|
| YDR119W-A | 1.20 |
Putative protein of unknown function |
||
| YHR096C | 1.20 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YMR180C | 1.19 |
CTL1
|
RNA 5'-triphosphatase, localizes to both the nucleus and cytoplasm |
|
| YGL045W | 1.19 |
RIM8
|
Protein of unknown function, involved in the proteolytic activation of Rim101p in response to alkaline pH; has similarity to A. nidulans PalF |
|
| YJR150C | 1.19 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YKL102C | 1.18 |
Dubious open reading frame unlikely to encode a functional protein; deletion confers sensitivity to citric acid; predicted protein would include a thiol-disulfide oxidoreductase active site |
||
| YMR191W | 1.17 |
SPG5
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YEL070W | 1.15 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YGR243W | 1.12 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YBL100C | 1.12 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the 5' end of ATP1 |
||
| YDR018C | 1.12 |
Probable membrane protein with three predicted transmembrane domains; homologous to Ybr042cp, similar to C. elegans F55A11.5 and maize 1-acyl-glycerol-3-phosphate acyltransferase; null exhibits no apparent phenotype |
||
| YPR192W | 1.10 |
AQY1
|
Spore-specific water channel that mediates the transport of water across cell membranes, developmentally controlled; may play a role in spore maturation, probably by allowing water outflow, may be involved in freeze tolerance |
|
| YMR068W | 1.10 |
AVO2
|
Component of a complex containing the Tor2p kinase and other proteins, which may have a role in regulation of cell growth |
|
| YNL144C | 1.09 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YDR199W | 1.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene VPS64; computationally predicted to have thiol-disulfide oxidoreductase activity |
||
| YGR022C | 1.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YOR100C | 1.09 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YMR256C | 1.08 |
COX7
|
Subunit VII of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YCR086W | 1.08 |
CSM1
|
Nucleolar protein that forms a complex with Lrs4p which binds Mam1p at kinetochores during meiosis I to mediate accurate chromosome segregation, may be involved in premeiotic DNA replication; possible role in telomere maintenance |
|
| YLR279W | 1.07 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR097C | 1.07 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YGR070W | 1.07 |
ROM1
|
GDP/GTP exchange protein (GEP) for Rho1p; mutations are synthetically lethal with mutations in rom2, which also encodes a GEP |
|
| YFL061W | 1.07 |
DDI2
|
Protein whose expression is induced by DNA damage |
|
| YJL133C-A | 1.07 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YLR308W | 1.05 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YAL063C | 1.05 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YBR072W | 1.05 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YJL089W | 1.05 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YDR406W | 1.05 |
PDR15
|
Plasma membrane ATP binding cassette (ABC) transporter, multidrug transporter and general stress response factor implicated in cellular detoxification; regulated by Pdr1p, Pdr3p and Pdr8p; promoter contains a PDR responsive element |
|
| YPR193C | 1.04 |
HPA2
|
Tetrameric histone acetyltransferase with similarity to Gcn5p, Hat1p, Elp3p, and Hpa3p; acetylates histones H3 and H4 in vitro and exhibits autoacetylation activity |
|
| YGL117W | 1.04 |
Putative protein of unknown function |
||
| YDR464W | 1.04 |
SPP41
|
Protein involved in negative regulation of expression of spliceosome components PRP4 and PRP3 |
|
| YGR230W | 1.04 |
BNS1
|
Protein with some similarity to Spo12p; overexpression bypasses need for Spo12p, but not required for meiosis |
|
| YEL009C | 1.04 |
GCN4
|
Transcriptional activator of amino acid biosynthetic genes in response to amino acid starvation; expression is tightly regulated at both the transcriptional and translational levels |
|
| YDL170W | 1.04 |
UGA3
|
Transcriptional activator necessary for gamma-aminobutyrate (GABA)-dependent induction of GABA genes (such as UGA1, UGA2, UGA4); zinc-finger transcription factor of the Zn(2)-Cys(6) binuclear cluster domain type; localized to the nucleus |
|
| YDL214C | 1.03 |
PRR2
|
Serine/threonine protein kinase that inhibits pheromone induced signalling downstream of MAPK, possibly at the level of the Ste12p transcription factor |
|
| YPR030W | 1.03 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YOR393W | 1.02 |
ERR1
|
Protein of unknown function, has similarity to enolases |
|
| YEL033W | 1.02 |
MTC7
|
Predicted metabolic role based on network analysis derived from ChIP experiments, a large-scale deletion study and localization of transcription factor binding sites; null mutant is sensitive to temperature oscillation in a cdc13-1 mutant |
|
| YFR010W-A | 1.02 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the uncharacterized gene YFR011C; identified by expression profiling and mass spectrometry |
||
| YCL048W | 1.02 |
SPS22
|
Protein of unknown function, redundant with Sps2p for the organization of the beta-glucan layer of the spore wall |
|
| YDR171W | 1.01 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YAR068W | 1.01 |
Fungal-specific protein of unknown function; induced in respiratory-deficient cells |
||
| YAL018C | 1.01 |
Putative protein of unknown function |
||
| YNL318C | 1.01 |
HXT14
|
Protein with similarity to hexose transporter family members, expression is induced in low glucose and repressed in high glucose; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YNL012W | 1.00 |
SPO1
|
Meiosis-specific protein with similarity to phospholipase B, required for meiotic spindle pole body duplication and separation; required for spore formation |
|
| YDR092W | 1.00 |
UBC13
|
Ubiquitin-conjugating enzyme involved in the error-free DNA postreplication repair pathway; interacts with Mms2p to assemble ubiquitin chains at the Ub Lys-63 residue; DNA damage triggers redistribution from the cytoplasm to the nucleus |
|
| YOR344C | 1.00 |
TYE7
|
Serine-rich protein that contains a basic-helix-loop-helix (bHLH) DNA binding motif; binds E-boxes of glycolytic genes and contributes to their activation; may function as a transcriptional activator in Ty1-mediated gene expression |
|
| YOR394W | 0.99 |
PAU21
|
Hypothetical protein |
|
| YOR388C | 0.99 |
FDH1
|
NAD(+)-dependent formate dehydrogenase, may protect cells from exogenous formate |
|
| YJR159W | 0.99 |
SOR1
|
Sorbitol dehydrogenase; expression is induced in the presence of sorbitol |
|
| YCL012C | 0.99 |
Putative protein of unknown function; orthologs are present in S. bayanus, S. paradoxus and Ashbya gossypii; YCL012C is not an essential gene |
||
| YGR023W | 0.99 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YFR023W | 0.98 |
PES4
|
Poly(A) binding protein, suppressor of DNA polymerase epsilon mutation, similar to Mip6p |
|
| YBR040W | 0.98 |
FIG1
|
Integral membrane protein required for efficient mating; may participate in or regulate the low affinity Ca2+ influx system, which affects intracellular signaling and cell-cell fusion during mating |
|
| YPR026W | 0.98 |
ATH1
|
Acid trehalase required for utilization of extracellular trehalose |
|
| YPR127W | 0.98 |
Putative protein of unknown function; expression is activated by transcription factor YRM1/YOR172W; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YIL160C | 0.98 |
POT1
|
3-ketoacyl-CoA thiolase with broad chain length specificity, cleaves 3-ketoacyl-CoA into acyl-CoA and acetyl-CoA during beta-oxidation of fatty acids |
|
| YCL001W-B | 0.98 |
Putative protein of unknown function; identified by homology |
||
| YKR073C | 0.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR431C | 0.97 |
ATG23
|
Peripheral membrane protein required for the cytoplasm-to-vacuole targeting (Cvt) pathway; cycles between the pre-autophagosome (PAS) and non-PAS locations; forms a complex with Atg9p and Atg27p |
|
| YBR230C | 0.97 |
OM14
|
Integral mitochondrial outer membrane protein; abundance is decreased in cells grown in glucose relative to other carbon sources; appears to contain 3 alpha-helical transmembrane segments; ORF encodes a 97-basepair intron |
|
| YIR014W | 0.97 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; expression directly regulated by the metabolic and meiotic transcriptional regulator Ume6p; YIR014W is a non-essential gene |
||
| YPR078C | 0.97 |
Putative protein of unknown function; possible role in DNA metabolism and/or in genome stability; expression is heat-inducible |
||
| YIL136W | 0.96 |
OM45
|
Protein of unknown function, major constituent of the mitochondrial outer membrane; located on the outer (cytosolic) face of the outer membrane |
|
| YDL172C | 0.96 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR251W | 0.95 |
GTO3
|
Omega class glutathione transferase; putative cytosolic localization |
|
| YNL018C | 0.95 |
Putative protein of unknown function |
||
| YBL048W | 0.94 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YGL230C | 0.94 |
Putative protein of unknown function; non-essential gene |
||
| YOR355W | 0.94 |
GDS1
|
Protein of unknown function, required for growth on glycerol as a carbon source; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YHL041W | 0.93 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YBR295W | 0.93 |
PCA1
|
Cadmium transporting P-type ATPase; may also have a role in copper and iron homeostasis; S288C and most other lab strains contain a G970R mutation which eliminates normal cadmium transport function |
|
| YLL029W | 0.93 |
FRA1
|
Protein involved in negative regulation of transcription of iron regulon; forms an iron independent complex with Fra2p, Grx3p, and Grx4p; cytosolic; mutant fails to repress transcription of iron regulon and is defective in spore formation |
|
| YJR154W | 0.93 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YKL038W | 0.92 |
RGT1
|
Glucose-responsive transcription factor that regulates expression of several glucose transporter (HXT) genes in response to glucose; binds to promoters and acts both as a transcriptional activator and repressor |
Network of associatons between targets according to the STRING database.
First level regulatory network of ORC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0019626 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 1.3 | 3.8 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 1.2 | 7.0 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 1.1 | 4.4 | GO:0006848 | pyruvate transport(GO:0006848) |
| 1.0 | 2.9 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.9 | 2.8 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.8 | 2.5 | GO:0009593 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.8 | 3.2 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.7 | 2.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.7 | 2.0 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.7 | 0.7 | GO:0001178 | regulation of transcriptional start site selection at RNA polymerase II promoter(GO:0001178) |
| 0.6 | 1.2 | GO:0035948 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.6 | 1.8 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.6 | 5.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.5 | 9.2 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.5 | 1.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 2.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.5 | 1.5 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.5 | 1.0 | GO:0045981 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.5 | 2.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.5 | 2.4 | GO:0006026 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.4 | 1.7 | GO:0046323 | glucose import(GO:0046323) |
| 0.4 | 3.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 7.4 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.4 | 2.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.4 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.4 | 2.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.4 | 1.2 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.4 | 0.4 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.4 | 1.2 | GO:0070583 | spore membrane bending pathway(GO:0070583) |
| 0.4 | 1.6 | GO:0034487 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.4 | 1.2 | GO:0006171 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.4 | 1.9 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.4 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 1.9 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.4 | 1.5 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.4 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.4 | 2.3 | GO:0090294 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.4 | 3.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 3.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.4 | 2.6 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.4 | 1.1 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.4 | 3.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.4 | 1.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.4 | 0.4 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.3 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 2.4 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.3 | 0.3 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.3 | 3.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 1.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) |
| 0.3 | 2.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.3 | 1.9 | GO:0001041 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.3 | 1.3 | GO:0031138 | negative regulation of conjugation(GO:0031135) negative regulation of conjugation with cellular fusion(GO:0031138) negative regulation of multi-organism process(GO:0043901) |
| 0.3 | 1.6 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.3 | 0.3 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.3 | 1.2 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.3 | 0.6 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 2.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.3 | 0.9 | GO:0015691 | cadmium ion transport(GO:0015691) |
| 0.3 | 4.2 | GO:0000767 | cell morphogenesis involved in conjugation(GO:0000767) |
| 0.3 | 1.6 | GO:0032120 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.3 | 1.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 1.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.3 | 1.6 | GO:0051322 | anaphase(GO:0051322) |
| 0.3 | 1.9 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 1.6 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.3 | 0.3 | GO:0018209 | peptidyl-serine phosphorylation(GO:0018105) peptidyl-serine modification(GO:0018209) |
| 0.3 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 0.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 1.0 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.3 | 2.3 | GO:0015688 | iron chelate transport(GO:0015688) |
| 0.3 | 1.3 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.3 | 2.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.3 | 1.5 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.2 | 0.7 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 1.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 0.9 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.2 | 1.6 | GO:0061013 | regulation of mRNA catabolic process(GO:0061013) |
| 0.2 | 0.7 | GO:0050810 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.2 | 2.4 | GO:0015893 | drug transport(GO:0015893) |
| 0.2 | 1.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.2 | 0.9 | GO:0000255 | allantoin metabolic process(GO:0000255) allantoin catabolic process(GO:0000256) cellular amide catabolic process(GO:0043605) |
| 0.2 | 1.3 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.0 | GO:0045033 | peroxisome inheritance(GO:0045033) |
| 0.2 | 1.0 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.2 | 0.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.2 | 0.6 | GO:0090295 | nitrogen catabolite repression of transcription(GO:0090295) |
| 0.2 | 1.4 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.2 | 0.6 | GO:0009300 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.2 | 0.6 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.2 | 2.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 0.6 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.2 | 0.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 2.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 0.6 | GO:0045128 | negative regulation of reciprocal meiotic recombination(GO:0045128) |
| 0.2 | 6.3 | GO:0010927 | cellular component assembly involved in morphogenesis(GO:0010927) |
| 0.2 | 0.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.2 | 1.4 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.2 | 0.9 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.2 | 0.5 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.2 | 0.5 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.2 | 1.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.2 | 0.7 | GO:0000755 | cytogamy(GO:0000755) |
| 0.2 | 0.7 | GO:0071331 | cellular glucose homeostasis(GO:0001678) carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 0.5 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.2 | 1.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.2 | 0.7 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.2 | 0.3 | GO:0000078 | obsolete cytokinesis after mitosis checkpoint(GO:0000078) |
| 0.2 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.2 | 1.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 0.6 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.2 | 0.5 | GO:1902749 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) mitotic G2/M transition checkpoint(GO:0044818) regulation of cell cycle G2/M phase transition(GO:1902749) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.2 | 0.6 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 2.5 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.2 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.2 | 0.5 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.2 | 1.1 | GO:0006641 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.2 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.8 | GO:0031032 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.2 | 0.3 | GO:0032874 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.1 | 0.7 | GO:0043112 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.1 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.4 | GO:1903339 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.1 | 2.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 3.8 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.1 | 0.3 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.1 | 1.0 | GO:0045931 | positive regulation of mitotic cell cycle(GO:0045931) |
| 0.1 | 0.3 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 0.1 | GO:0001323 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.6 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.1 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.6 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.3 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.1 | 0.2 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.6 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.2 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.1 | 0.5 | GO:0001308 | negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.1 | 2.1 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 0.8 | GO:0009231 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.1 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.3 | GO:0006003 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.3 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.7 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.1 | 1.0 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 0.2 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.1 | 0.5 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 1.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.4 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.1 | 0.3 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.1 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.4 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 1.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.5 | GO:0006476 | protein deacetylation(GO:0006476) protein deacylation(GO:0035601) macromolecule deacylation(GO:0098732) |
| 0.1 | 0.4 | GO:0006078 | (1->6)-beta-D-glucan biosynthetic process(GO:0006078) |
| 0.1 | 0.1 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.1 | GO:0044109 | cellular alcohol catabolic process(GO:0044109) |
| 0.1 | 2.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.2 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.2 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) arabinose catabolic process(GO:0019568) |
| 0.1 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 0.8 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.1 | 0.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) re-entry into mitotic cell cycle after pheromone arrest(GO:0000321) |
| 0.1 | 0.3 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.1 | 0.4 | GO:0031930 | mitochondria-nucleus signaling pathway(GO:0031930) |
| 0.1 | 0.3 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 0.8 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.7 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.3 | GO:0001934 | positive regulation of protein phosphorylation(GO:0001934) |
| 0.1 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.1 | 1.0 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.1 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.3 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.4 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.6 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 0.9 | GO:0042775 | respiratory electron transport chain(GO:0022904) ATP synthesis coupled electron transport(GO:0042773) mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.1 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.1 | 0.1 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) protein-DNA complex remodeling(GO:0001120) macromolecular complex remodeling(GO:0034367) |
| 0.1 | 0.7 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.1 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.5 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.1 | GO:0099515 | actin filament-based movement(GO:0030048) actin filament-based transport(GO:0099515) |
| 0.1 | 0.2 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 3.5 | GO:0048646 | anatomical structure formation involved in morphogenesis(GO:0048646) |
| 0.1 | 0.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.6 | GO:0000902 | cell morphogenesis(GO:0000902) |
| 0.1 | 0.5 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.4 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.1 | GO:0000087 | mitotic M phase(GO:0000087) M phase(GO:0000279) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.0 | GO:0051403 | stress-activated protein kinase signaling cascade(GO:0031098) regulation of stress-activated MAPK cascade(GO:0032872) stress-activated MAPK cascade(GO:0051403) regulation of stress-activated protein kinase signaling cascade(GO:0070302) |
| 0.0 | 0.1 | GO:0070054 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 | 0.4 | GO:0009262 | deoxyribonucleotide metabolic process(GO:0009262) deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0045005 | DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0018195 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0051346 | negative regulation of hydrolase activity(GO:0051346) |
| 0.0 | 0.2 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.0 | GO:1902533 | positive regulation of intracellular signal transduction(GO:1902533) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.4 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.0 | GO:0009967 | positive regulation of signal transduction(GO:0009967) positive regulation of signaling(GO:0023056) |
| 0.0 | 0.2 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0051156 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0006077 | (1->6)-beta-D-glucan metabolic process(GO:0006077) |
| 0.0 | 0.0 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.0 | 0.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.1 | GO:0019320 | hexose catabolic process(GO:0019320) |
| 0.0 | 0.1 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 0.0 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.2 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.0 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.1 | GO:0045833 | negative regulation of lipid metabolic process(GO:0045833) |
| 0.0 | 0.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.0 | 0.9 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0007532 | regulation of mating-type specific transcription, DNA-templated(GO:0007532) |
| 0.0 | 0.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.0 | GO:2000218 | regulation of invasive growth in response to glucose limitation(GO:2000217) negative regulation of invasive growth in response to glucose limitation(GO:2000218) |
| 0.0 | 0.0 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.0 | 0.1 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.1 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.0 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0044426 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.5 | 1.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.5 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.4 | 1.3 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.4 | 0.9 | GO:0001400 | mating projection base(GO:0001400) |
| 0.4 | 4.2 | GO:0031160 | spore wall(GO:0031160) |
| 0.3 | 1.4 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.3 | 1.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.3 | 2.2 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.3 | 1.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 7.0 | GO:0042763 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.3 | 2.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 0.7 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.2 | 0.7 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 3.3 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.2 | 0.7 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.2 | 1.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 0.9 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.2 | 0.8 | GO:0030287 | cell wall-bounded periplasmic space(GO:0030287) |
| 0.2 | 3.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.2 | 0.7 | GO:0008623 | CHRAC(GO:0008623) |
| 0.2 | 1.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 4.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.9 | GO:0000133 | polarisome(GO:0000133) |
| 0.1 | 0.6 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.4 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.1 | 0.7 | GO:0044233 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.6 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.1 | 0.5 | GO:0030892 | mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) |
| 0.1 | 4.9 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 3.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.5 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.1 | 0.5 | GO:0000799 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.1 | 0.7 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.3 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 3.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.5 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.5 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 2.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 3.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 0.2 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.1 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.8 | GO:0033698 | Rpd3L complex(GO:0033698) |
| 0.0 | 4.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.2 | GO:0045334 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0070274 | RES complex(GO:0070274) |
| 0.0 | 1.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.9 | GO:0005937 | mating projection(GO:0005937) cell projection(GO:0042995) |
| 0.0 | 0.2 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0032176 | split septin rings(GO:0032176) cellular bud neck split septin rings(GO:0032177) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 3.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0005824 | outer plaque of spindle pole body(GO:0005824) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0098562 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0097525 | spliceosomal snRNP complex(GO:0097525) |
| 0.0 | 0.1 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 0.0 | GO:0051286 | cell tip(GO:0051286) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 1.5 | 6.1 | GO:0005537 | mannose binding(GO:0005537) |
| 1.2 | 6.0 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.9 | 2.8 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.9 | 3.7 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.8 | 3.4 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.8 | 2.5 | GO:0005536 | glucose binding(GO:0005536) |
| 0.8 | 3.2 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.8 | 3.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.6 | 1.8 | GO:0016208 | AMP binding(GO:0016208) |
| 0.6 | 3.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.6 | 1.8 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.6 | 2.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.6 | 1.7 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 2.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.5 | 3.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.5 | 1.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.4 | 2.6 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.4 | 1.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.4 | 1.6 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.4 | 1.9 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.3 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 3.4 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.3 | 1.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 1.0 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.3 | 1.3 | GO:0016725 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 2.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 1.9 | GO:0001003 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.3 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.4 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 0.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.3 | 1.4 | GO:0050253 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 1.1 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.3 | 0.8 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.3 | 5.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.3 | 0.3 | GO:0019902 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.3 | 2.7 | GO:0015145 | monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.3 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 1.0 | GO:0000772 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.3 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 1.0 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.2 | 1.0 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.2 | 4.1 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.2 | 0.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.2 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 1.3 | GO:0008106 | aldo-keto reductase (NADP) activity(GO:0004033) alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.2 | 2.4 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.2 | 0.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.2 | 2.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.2 | 0.8 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.2 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 5.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 3.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 0.7 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.2 | 2.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 0.7 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 1.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.5 | GO:0016898 | lactate dehydrogenase activity(GO:0004457) D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.2 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.6 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 0.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 1.2 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 1.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 3.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 5.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 0.1 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 2.8 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.1 | 0.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 1.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0016878 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.1 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.1 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 0.7 | GO:0038023 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.1 | 0.4 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.1 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.9 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.1 | GO:0001007 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) |
| 0.1 | 0.6 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.1 | 6.0 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.1 | 1.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.2 | GO:0016642 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.1 | 0.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.1 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.2 | GO:0048256 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.1 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0001128 | RNA polymerase II transcription coactivator activity involved in preinitiation complex assembly(GO:0001128) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.0 | 0.1 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.0 | 2.3 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.0 | 0.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.0 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.0 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.0 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.2 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.2 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 106.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.0 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.8 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 2.8 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.2 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 0.2 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.1 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 105.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME RNA POL II TRANSCRIPTION | Genes involved in RNA Polymerase II Transcription |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME MEIOSIS | Genes involved in Meiosis |