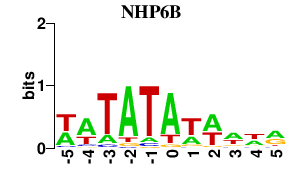
Results for NHP6B
Z-value: 0.64
Transcription factors associated with NHP6B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHP6B
|
S000002157 | High-mobility group (HMG) protein, binds to and remodels nucleosomes |
Activity-expression correlation:
Activity profile of NHP6B motif
Sorted Z-values of NHP6B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YPR169W-A | 2.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YKR034W | 1.90 |
DAL80
|
Negative regulator of genes in multiple nitrogen degradation pathways; expression is regulated by nitrogen levels and by Gln3p; member of the GATA-binding family, forms homodimers and heterodimers with Deh1p |
|
| YHR126C | 1.11 |
Putative protein of unknown function; transcription dependent upon Azf1p |
||
| YKR033C | 1.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene DAL80 |
||
| YFL019C | 1.02 |
Dubious open reading frame unlikely to encode a protein; YFL019C is not an essential gene |
||
| YMR107W | 1.01 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YHR079C-A | 1.01 |
SAE3
|
Meiosis specific protein involved in DMC1-dependent meiotic recombination, forms heterodimer with Mei5p; proposed to be an assembly factor for Dmc1p |
|
| YKL221W | 0.99 |
MCH2
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YBR292C | 0.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YPL026C | 0.92 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YKL065W-A | 0.83 |
Putative protein of unknown function |
||
| YJL149W | 0.82 |
DAS1
|
Putative SCF ubiquitin ligase F-box protein; interacts physically with both Cdc53p and Skp1 and genetically with CDC34; similar to putative F-box protein YDR131C; null mutant suppresses dst1delta sensitivity for 6-azauracil |
|
| YLR307C-A | 0.81 |
Putative protein of unknown function |
||
| YJR094C | 0.81 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YDL210W | 0.80 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YFL020C | 0.77 |
PAU5
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YDR446W | 0.69 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YMR193C-A | 0.68 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJL205C | 0.66 |
NCE101
|
Protein of unknown function, involved in secretion of proteins that lack classical secretory signal sequences |
|
| YHR096C | 0.66 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YBR240C | 0.65 |
THI2
|
Zinc finger protein of the Zn(II)2Cys6 type, probable transcriptional activator of thiamine biosynthetic genes |
|
| YPL187W | 0.63 |
MF(ALPHA)1
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)2, although MF(ALPHA)1 produces most alpha-factor |
|
| YGR039W | 0.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YNL014W | 0.61 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YLR308W | 0.61 |
CDA2
|
Chitin deacetylase, together with Cda1p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YPL222C-A | 0.60 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YGL227W | 0.58 |
VID30
|
Protein involved in proteasome-dependent catabolite degradation of fructose-1,6-bisphosphatase (FBPase); shifts the balance of nitrogen metabolism toward the production of glutamate; localized to the nucleus and the cytoplasm |
|
| YIL045W | 0.56 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YML047C | 0.56 |
PRM6
|
Pheromone-regulated protein, predicted to have 2 transmembrane segments; regulated by Ste12p during mating |
|
| YAL062W | 0.56 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YOL045W | 0.56 |
PSK2
|
One of two (see also PSK1) PAS domain containing S/T protein kinases; regulates sugar flux and translation in response to an unknown metabolite by phosphorylating Ugp1p and Gsy2p (sugar flux) and Caf20p, Tif11p and Sro9p (translation) |
|
| YKL123W | 0.55 |
Dubious open reading frame, unlikely to encode a protein; partially overlaps the verified gene SSH4 |
||
| YBR076C-A | 0.55 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YBR157C | 0.55 |
ICS2
|
Protein of unknown function; null mutation does not confer any obvious defects in growth, spore germination, viability, or carbohydrate utilization |
|
| YOL118C | 0.54 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YPL156C | 0.53 |
PRM4
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; transcriptionally regulated by Ste12p during mating and by Cat8p during the diauxic shift |
|
| YMR194C-B | 0.52 |
Putative protein of unknown function |
||
| YLL046C | 0.52 |
RNP1
|
Ribonucleoprotein that contains two RNA recognition motifs (RRM) |
|
| YDL138W | 0.52 |
RGT2
|
Plasma membrane glucose receptor, highly similar to Snf3p; both Rgt2p and Snf3p serve as transmembrane glucose sensors generating an intracellular signal that induces expression of glucose transporter (HXT) genes |
|
| YCL001W-B | 0.51 |
Putative protein of unknown function; identified by homology |
||
| YJL150W | 0.51 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YCR007C | 0.51 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YLL013C | 0.50 |
PUF3
|
Protein of the mitochondrial outer surface, links the Arp2/3 complex with the mitochore during anterograde mitochondrial movement; also binds to and promotes degradation of mRNAs for select nuclear-encoded mitochondrial proteins |
|
| YEL014C | 0.50 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPR001W | 0.49 |
CIT3
|
Dual specificity mitochondrial citrate and methylcitrate synthase; catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate and that of propionyl-CoA and oxaloacetate to form 2-methylcitrate |
|
| YBL033C | 0.48 |
RIB1
|
GTP cyclohydrolase II; catalyzes the first step of the riboflavin biosynthesis pathway |
|
| YMR090W | 0.48 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YDL139C | 0.47 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YDR445C | 0.47 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL211C | 0.46 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YLR296W | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR171W | 0.46 |
BSP1
|
Adapter that links synaptojanins Inp52p and Inp53p to the cortical actin cytoskeleton |
|
| YKL109W | 0.45 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YPL024W | 0.45 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YGL177W | 0.45 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER167W | 0.45 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YDL223C | 0.44 |
HBT1
|
Substrate of the Hub1p ubiquitin-like protein that localizes to the shmoo tip (mating projection); mutants are defective for mating projection formation, thereby implicating Hbt1p in polarized cell morphogenesis |
|
| YMR104C | 0.44 |
YPK2
|
Protein kinase with similarityto serine/threonine protein kinase Ypk1p; functionally redundant with YPK1 at the genetic level; participates in a signaling pathway required for optimal cell wall integrity; homolog of mammalian kinase SGK |
|
| YDR504C | 0.43 |
SPG3
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YHR217C | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; located in the telomeric region TEL08R. |
||
| YOL034W | 0.43 |
SMC5
|
Structural maintenance of chromosomes (SMC) protein; essential subunit of the Mms21-Smc5-Smc6 complex; required for growth and DNA repair; S. pombe homolog forms a heterodimer with S. pombe Rad18p that is involved in DNA repair |
|
| YMR195W | 0.43 |
ICY1
|
Protein of unknown function, required for viability in rich media of cells lacking mitochondrial DNA; mutants have an invasive growth defect with elongated morphology; induced by amino acid starvation |
|
| YML047W-A | 0.42 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR056W | 0.42 |
RSC1
|
Component of the RSC chromatin remodeling complex; required for expression of mid-late sporulation-specific genes; contains two essential bromodomains, a bromo-adjacent homology (BAH) domain, and an AT hook |
|
| YKL177W | 0.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene STE3 |
||
| YPR193C | 0.40 |
HPA2
|
Tetrameric histone acetyltransferase with similarity to Gcn5p, Hat1p, Elp3p, and Hpa3p; acetylates histones H3 and H4 in vitro and exhibits autoacetylation activity |
|
| YNL180C | 0.40 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YNL179C | 0.40 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YOR062C | 0.40 |
Protein of unknown function; similar to YKR075Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YLR124W | 0.40 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL105C | 0.40 |
Putative protein of unknown function |
||
| YDL130W-A | 0.39 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YMR204C | 0.39 |
INP1
|
Peripheral membrane protein of peroxisomes involved in peroxisomal inheritance |
|
| YOR192C-C | 0.39 |
Putative protein of unknown function; identified by expression profiling and mass spectrometry |
||
| YEL007W | 0.37 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YBL067C | 0.37 |
UBP13
|
Putative ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YBR296C | 0.37 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YBL097W | 0.37 |
BRN1
|
Essential protein required for chromosome condensation, likely to function as an intrinsic component of the condensation machinery, may influence multiple aspects of chromosome transmission and dynamics |
|
| YKR032W | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGL117W | 0.36 |
Putative protein of unknown function |
||
| YLR408C | 0.36 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endosome; YLR408C is not an essential gene |
||
| YCR001W | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YCR001W is not an essential gene |
||
| YAR069C | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR467C | 0.36 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR307W | 0.35 |
CDA1
|
Chitin deacetylase, together with Cda2p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YJL116C | 0.35 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YLR327C | 0.35 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YHR095W | 0.35 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YCL012C | 0.35 |
Putative protein of unknown function; orthologs are present in S. bayanus, S. paradoxus and Ashbya gossypii; YCL012C is not an essential gene |
||
| YOL159C-A | 0.35 |
Putative protein of unknown function; identified by sequence comparison with hemiascomycetous yeast species |
||
| YPL020C | 0.35 |
ULP1
|
Ubl (ubiquitin-like protein)-specific protease that cleaves Smt3p protein conjugates; specifically required for cell cycle progression; associates with nucleoporins and may interact with septin rings during telophase |
|
| YLR309C | 0.35 |
IMH1
|
Protein involved in vesicular transport, mediates transport between an endosomal compartment and the Golgi, contains a Golgi-localization (GRIP) domain that interacts with activated Arl1p-GTP to localize Imh1p to the Golgi |
|
| YKL178C | 0.34 |
STE3
|
Receptor for a factor receptor, transcribed in alpha cells and required for mating by alpha cells, couples to MAP kinase cascade to mediate pheromone response; ligand bound receptors are endocytosed and recycled to the plasma membrane; GPCR |
|
| YLL019C | 0.34 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YKL188C | 0.33 |
PXA2
|
Subunit of a heterodimeric peroxisomal ATP-binding cassette transporter complex (Pxa1p-Pxa2p), required for import of long-chain fatty acids into peroxisomes; similarity to human adrenoleukodystrophy transporter and ALD-related proteins |
|
| YLR357W | 0.33 |
RSC2
|
Component of the RSC chromatin remodeling complex; required for expression of mid-late sporulation-specific genes; involved in telomere maintenance |
|
| YML082W | 0.33 |
Putative protein predicted to have carbon-sulfur lyase activity; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and the cytoplasm; YNML082W is not an essential gene |
||
| YER121W | 0.33 |
Putative protein of unknown function; may be involved in phosphatase regulation and/or generation of precursor metabolites and energy |
||
| YDR022C | 0.33 |
CIS1
|
Autophagy-specific protein required for autophagosome formation; may form a complex with Atg17p and Atg29p that localizes other proteins to the pre-autophagosomal structure; high-copy suppressor of CIK1 deletion |
|
| YKR039W | 0.33 |
GAP1
|
General amino acid permease; localization to the plasma membrane is regulated by nitrogen source |
|
| YGR226C | 0.33 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; overlaps significantly with a verified ORF, AMA1/YGR225W |
||
| YAL066W | 0.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR140W | 0.32 |
SFL1
|
Transcriptional repressor and activator; involved in repression of flocculation-related genes, and activation of stress responsive genes; negatively regulated by cAMP-dependent protein kinase A subunit Tpk2p |
|
| YPL258C | 0.32 |
THI21
|
Hydroxymethylpyrimidine phosphate kinase, involved in the last steps in thiamine biosynthesis; member of a gene family with THI20 and THI22; functionally redundant with Thi20p |
|
| YOR139C | 0.32 |
Hypothetical protein |
||
| YPR120C | 0.32 |
CLB5
|
B-type cyclin involved in DNA replication during S phase; activates Cdc28p to promote initiation of DNA synthesis; functions in formation of mitotic spindles along with Clb3p and Clb4p; most abundant during late G1 phase |
|
| YER097W | 0.32 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL197W | 0.32 |
MDS3
|
Protein with an N-terminal kelch-like domain, putative negative regulator of early meiotic gene expression; required, with Pmd1p, for growth under alkaline conditions |
|
| YGR239C | 0.32 |
PEX21
|
Peroxin required for targeting of peroxisomal matrix proteins containing PTS2; interacts with Pex7p; partially redundant with Pex18p |
|
| YML068W | 0.32 |
ITT1
|
Protein that modulates the efficiency of translation termination, interacts with translation release factors eRF1 (Sup45p) and eRF3 (Sup35p) in vitro, contains a zinc finger domain characteristic of the TRIAD class of proteins |
|
| YAR070C | 0.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLL025W | 0.31 |
PAU17
|
Putative protein of unknown function; YLL025W is not an essential gene |
|
| YOL035C | 0.31 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOR011W-A | 0.31 |
Putative protein of unknown function |
||
| YHR030C | 0.31 |
SLT2
|
Serine/threonine MAP kinase involved in regulating the maintenance of cell wall integrity and progression through the cell cycle; regulated by the PKC1-mediated signaling pathway |
|
| YDL028C | 0.31 |
MPS1
|
Dual-specificity kinase required for spindle pole body (SPB) duplication and spindle checkpoint function; substrates include SPB proteins Spc42p, Spc110p, and Spc98p, mitotic exit network protein Mob1p, and checkpoint protein Mad1p |
|
| YBL053W | 0.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR032W | 0.30 |
GSC2
|
Catalytic subunit of 1,3-beta-glucan synthase, involved in formation of the inner layer of the spore wall; activity positively regulated by Rho1p and negatively by Smk1p; has similarity to an alternate catalytic subunit, Fks1p (Gsc1p) |
|
| YAL067C | 0.30 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YGR286C | 0.30 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YFL033C | 0.30 |
RIM15
|
Glucose-repressible protein kinase involved in signal transduction during cell proliferation in response to nutrients, specifically the establishment of stationary phase; identified as a regulator of IME2; substrate of Pho80p-Pho85p kinase |
|
| YKR009C | 0.30 |
FOX2
|
Multifunctional enzyme of the peroxisomal fatty acid beta-oxidation pathway; has 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase activities |
|
| YER187W | 0.29 |
Putative protein of unknown function; induced in respiratory-deficient cells |
||
| YJR128W | 0.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YGL170C | 0.29 |
SPO74
|
Component of the meiotic outer plaque of the spindle pole body, involved in modifying the meiotic outer plaque that is required prior to prospore membrane formation |
|
| YDR119W-A | 0.29 |
Putative protein of unknown function |
||
| YMR118C | 0.28 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YOR306C | 0.28 |
MCH5
|
Plasma membrane riboflavin transporter; facilitates the uptake of vitamin B2; required for FAD-dependent processes; sequence similarity to mammalian monocarboxylate permeases, however mutants are not deficient in monocarboxylate transport |
|
| YJR048W | 0.28 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YOR192C | 0.28 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YJL135W | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified genes YJL134W/LCB3 |
||
| YHR205W | 0.28 |
SCH9
|
Protein kinase involved in transcriptional activation of osmostress-responsive genes; regulates G1 progression, cAPK activity, nitrogen activation of the FGM pathway; involved in life span regulation; homologous to mammalian Akt/PKB |
|
| YLR092W | 0.27 |
SUL2
|
High affinity sulfate permease; sulfate uptake is mediated by specific sulfate transporters Sul1p and Sul2p, which control the concentration of endogenous activated sulfate intermediates |
|
| YCR066W | 0.27 |
RAD18
|
Protein involved in postreplication repair; binds single-stranded DNA and has single-stranded DNA dependent ATPase activity; forms heterodimer with Rad6p; contains RING-finger motif |
|
| YFL030W | 0.27 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YER054C | 0.27 |
GIP2
|
Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism; contains a conserved motif (GVNK motif) that is also found in Gac1p, Pig1p, and Pig2p |
|
| YGR087C | 0.27 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YAL063C | 0.27 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YFL058W | 0.27 |
THI5
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YDR389W | 0.26 |
SAC7
|
GTPase activating protein (GAP) for Rho1p, involved in signaling to the actin cytoskeleton, null mutations suppress tor2 mutations and temperature sensitive mutations in actin; potential Cdc28p substrate |
|
| YDR369C | 0.26 |
XRS2
|
Protein required for DNA repair; component of the Mre11 complex, which is involved in double strand breaks, meiotic recombination, telomere maintenance, and checkpoint signaling |
|
| YKL110C | 0.26 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YER129W | 0.26 |
SAK1
|
Upstream serine/threonine kinase for the SNF1 complex; partially redundant with Elm1p and Tos3p; members of this family have functional orthology with LKB1, a mammalian kinase associated with Peutz-Jeghers cancer-susceptibility syndrome |
|
| YHR071W | 0.26 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YER158W-A | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YDR223W | 0.26 |
CRF1
|
Transcriptional corepressor involved in the regulation of ribosomal protein gene transcription via the TOR signaling pathway and protein kinase A, phosphorylated by activated Yak1p which promotes accumulation of Crf1p in the nucleus |
|
| YGR161C | 0.25 |
RTS3
|
Putative component of the protein phosphatase type 2A complex |
|
| YGL096W | 0.25 |
TOS8
|
Homeodomain-containing transcription factor; SBF regulated target gene that in turn regulates expression of genes involved in G1/S phase events such as bud site selection, bud emergence and cell cycle progression; similarity to Cup9p |
|
| YFL036W | 0.25 |
RPO41
|
Mitochondrial RNA polymerase; single subunit enzyme similar to those of T3 and T7 bacteriophages; requires a specificity subunit encoded by MTF1 for promoter recognition |
|
| YDL085W | 0.25 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YPR006C | 0.25 |
ICL2
|
2-methylisocitrate lyase of the mitochondrial matrix, functions in the methylcitrate cycle to catalyze the conversion of 2-methylisocitrate to succinate and pyruvate; ICL2 transcription is repressed by glucose and induced by ethanol |
|
| YKL001C | 0.25 |
MET14
|
Adenylylsulfate kinase, required for sulfate assimilation and involved in methionine metabolism |
|
| YDL035C | 0.25 |
GPR1
|
Plasma membrane G protein coupled receptor (GPCR) that interacts with the heterotrimeric G protein alpha subunit, Gpa2p, and with Plc1p; sensor that integrates nutritional signals with the modulation of cell fate via PKA and cAMP synthesis |
|
| YBR184W | 0.25 |
Putative protein of unknown function; YBR184W is not an essential gene |
||
| YOR178C | 0.25 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YDR406W-A | 0.25 |
Dubious open reading frame unlikely to encode a functional protein; identified by fungal homology and RT-PCR |
||
| YDR392W | 0.24 |
SPT3
|
Subunit of the SAGA and SAGA-like transcriptional regulatory complexes, interacts with Spt15p to activate transcription of some RNA polymerase II-dependent genes, also functions to inhibit transcription at some promoters |
|
| YOL113W | 0.24 |
SKM1
|
Member of the PAK family of serine/threonine protein kinases with similarity to Ste20p and Cla4p; proposed to be a downstream effector of Cdc42p during polarized growth |
|
| YCL069W | 0.24 |
VBA3
|
Permease of basic amino acids in the vacuolar membrane |
|
| YGL089C | 0.24 |
MF(ALPHA)2
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)1, which is more highly expressed than MF(ALPHA)2 |
|
| YDR042C | 0.24 |
Putative protein of unknown function; expression is increased in ssu72-ts69 mutant |
||
| YKR105C | 0.24 |
VBA5
|
Putative transporter of the Major Facilitator Superfamily (MFS); proposed role as a basic amino acid permease based on phylogeny |
|
| YHR164C | 0.24 |
DNA2
|
Essential tripartite DNA replication factor with single-stranded DNA-dependent ATPase, ATP-dependent nuclease, and helicase activities; required for Okazaki fragment processing; involved in DNA repair pathways; potential Cdc28p substrate |
|
| YGR048W | 0.24 |
UFD1
|
Protein that interacts with Cdc48p and Npl4p, involved in recognition of polyubiquitinated proteins and their presentation to the 26S proteasome for degradation; involved in transporting proteins from the ER to the cytosol |
|
| YBR242W | 0.23 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YBR242W is not an essential gene |
||
| YDL246C | 0.23 |
SOR2
|
Protein of unknown function, computational analysis of large-scale protein-protein interaction data suggests a possible role in fructose or mannose metabolism |
|
| YKL106W | 0.23 |
AAT1
|
Mitochondrial aspartate aminotransferase, catalyzes the conversion of oxaloacetate to aspartate in aspartate and asparagine biosynthesis |
|
| YIL071C | 0.23 |
PCI8
|
Possible shared subunit of Cop9 signalosome (CSN) and eIF3, binds eIF3b subunit Prt1p, has possible dual functions in transcriptional and translational control, contains a PCI (Proteasome-COP9 signalosome (CSN)-eIF3) domain |
|
| YBR291C | 0.23 |
CTP1
|
Mitochondrial inner membrane citrate transporter, member of the mitochondrial carrier family |
|
| YOL051W | 0.23 |
GAL11
|
Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; affects transcription by acting as target of activators and repressors |
|
| YKL171W | 0.23 |
Putative protein of unknown function; predicted protein kinase; implicated in proteasome function; epitope-tagged protein localizes to the cytoplasm |
||
| YDL027C | 0.23 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YDL027C is not an essential gene |
||
| YOR329W-A | 0.23 |
Dubious open reading frame, unlikely to encode a functional protein; identified by fungal homology and RT-PCR |
||
| YNL171C | 0.23 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YGL036W | 0.22 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YGL036W is not an essential gene |
||
| YER014C-A | 0.22 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YPR065W | 0.22 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YKL066W | 0.22 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene YNK1 |
||
| YJL200C | 0.22 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YCL048W | 0.22 |
SPS22
|
Protein of unknown function, redundant with Sps2p for the organization of the beta-glucan layer of the spore wall |
|
| YGL118C | 0.22 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL027W | 0.22 |
SMA1
|
Protein of unknown function involved in the assembly of the prospore membrane during sporulation |
|
| YOL112W | 0.22 |
MSB4
|
GTPase-activating protein of the Ras superfamily that acts primarily on Sec4p, localizes to the bud site and bud tip, has similarity to Msb3p; msb3 msb4 double mutation causes defects in secretion and actin organization |
|
| YDL026W | 0.22 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER064C | 0.22 |
Non-essential nuclear protein; null mutation has global effects on transcription |
||
| YNL103W | 0.22 |
MET4
|
Leucine-zipper transcriptional activator, responsible for the regulation of the sulfur amino acid pathway, requires different combinations of the auxiliary factors Cbf1p, Met28p, Met31p and Met32p |
|
| YKR031C | 0.22 |
SPO14
|
Phospholipase D, catalyzes the hydrolysis of phosphatidylcholine, producing choline and phosphatidic acid; involved in Sec14p-independent secretion; required for meiosis and spore formation; differently regulated in secretion and meiosis |
|
| YNL036W | 0.22 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YGR072W | 0.22 |
UPF3
|
Component of the nonsense-mediated mRNA decay (NMD) pathway, along with Nam7p and Nmd2p; involved in decay of mRNA containing nonsense codons; involved in telomere maintenance |
|
| YKL076C | 0.21 |
PSY1
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 69% of ORF overlaps the uncharacterized ORF YKL075C |
|
| YPL034W | 0.21 |
Putative protein of unknown function; YPL034W is not essential gene |
||
| YNL267W | 0.21 |
PIK1
|
Phosphatidylinositol 4-kinase; catalyzes first step in the biosynthesis of phosphatidylinositol-4,5-biphosphate; may control cytokineses through the actin cytoskeleton |
|
| YLR337C | 0.21 |
VRP1
|
Proline-rich actin-associated protein involved in cytoskeletal organization and cytokinesis; related to mammalian Wiskott-Aldrich syndrome protein (WASP)-interacting protein (WIP) |
|
| YLL012W | 0.21 |
YEH1
|
Steryl ester hydrolase, one of three gene products (Yeh1p, Yeh2p, Tgl1p) responsible for steryl ester hydrolase activity and involved in sterol homeostasis; localized to lipid particle membranes |
|
| YEL045C | 0.21 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; deletion gives MMS sensitivity, growth defect under alkaline conditions, less than optimal growth upon citric acid stress |
||
| YDR043C | 0.21 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YDR421W | 0.21 |
ARO80
|
Zinc finger transcriptional activator of the Zn2Cys6 family; activates transcription of aromatic amino acid catabolic genes in the presence of aromatic amino acids |
|
| YOL117W | 0.21 |
RRI2
|
Subunit of the COP9 signalosome (CSN) complex that cleaves the ubiquitin-like protein Nedd8 from SCF ubiquitin ligases; plays a role in the mating pheromone response |
|
| YDL007C-A | 0.21 |
Putative protein of unknown function |
||
| YIL057C | 0.21 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
Network of associatons between targets according to the STRING database.
First level regulatory network of NHP6B
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0090295 | nitrogen catabolite repression of transcription(GO:0090295) |
| 0.3 | 0.8 | GO:0051594 | detection of chemical stimulus(GO:0009593) detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) detection of stimulus(GO:0051606) |
| 0.2 | 0.7 | GO:0019629 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.2 | 0.6 | GO:0070623 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.2 | 1.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) DNA recombinase assembly(GO:0000730) |
| 0.2 | 0.2 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.2 | 0.7 | GO:0042173 | regulation of sporulation resulting in formation of a cellular spore(GO:0042173) |
| 0.2 | 0.5 | GO:1900460 | negative regulation of invasive growth in response to glucose limitation by negative regulation of transcription from RNA polymerase II promoter(GO:1900460) |
| 0.2 | 0.5 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 0.7 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.2 | 0.5 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 0.8 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.1 | 0.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.8 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.9 | GO:0034284 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 1.2 | GO:0070873 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.1 | 0.3 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.1 | 0.7 | GO:0001323 | age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) |
| 0.1 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.6 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 | 0.3 | GO:0000304 | response to singlet oxygen(GO:0000304) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0045895 | positive regulation of mating-type specific transcription, DNA-templated(GO:0045895) |
| 0.1 | 0.4 | GO:0042357 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 0.4 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 | 0.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0006032 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.6 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.1 | 0.4 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.3 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.3 | GO:0045894 | regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001196) negative regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001198) negative regulation of mating-type specific transcription, DNA-templated(GO:0045894) |
| 0.1 | 0.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:0000755 | cytogamy(GO:0000755) |
| 0.1 | 0.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.3 | GO:0036498 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.1 | 0.4 | GO:0045033 | peroxisome inheritance(GO:0045033) |
| 0.1 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.2 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.2 | GO:0043937 | regulation of sporulation(GO:0043937) |
| 0.1 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) |
| 0.1 | 0.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 0.3 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) |
| 0.1 | 0.5 | GO:0009306 | protein secretion(GO:0009306) |
| 0.1 | 0.2 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.1 | 0.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.7 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.1 | 0.4 | GO:0070058 | tRNA gene clustering(GO:0070058) |
| 0.1 | 0.2 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.1 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.8 | GO:0000767 | cell morphogenesis involved in conjugation with cellular fusion(GO:0000753) cell morphogenesis involved in conjugation(GO:0000767) |
| 0.1 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.1 | 0.4 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.1 | GO:1903322 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.1 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.2 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.1 | 0.2 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.1 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.1 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of mitotic nuclear division(GO:0045840) positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.1 | 0.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.3 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.1 | GO:0005993 | trehalose catabolic process(GO:0005993) |
| 0.0 | 0.2 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) rDNA condensation(GO:0070550) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.3 | GO:0006276 | plasmid maintenance(GO:0006276) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) |
| 0.0 | 0.4 | GO:0070193 | synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.0 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.3 | GO:0001009 | transcription from RNA polymerase III type 2 promoter(GO:0001009) transcription from a RNA polymerase III hybrid type promoter(GO:0001041) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.0 | 0.4 | GO:0071709 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.1 | GO:0036095 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.0 | 0.2 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.0 | 0.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.6 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.2 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.0 | 0.0 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) positive regulation of cellular protein localization(GO:1903829) |
| 0.0 | 0.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.8 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1904357 | snoRNA transcription from an RNA polymerase II promoter(GO:0001015) negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.2 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0042744 | hydrogen peroxide metabolic process(GO:0042743) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.0 | 0.0 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0061393 | positive regulation of transcription from RNA polymerase II promoter in response to osmotic stress(GO:0061393) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) |
| 0.0 | 0.1 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.2 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.0 | 0.0 | GO:0071168 | establishment of protein localization to chromosome(GO:0070199) protein localization to chromatin(GO:0071168) establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.2 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:0071241 | cellular response to inorganic substance(GO:0071241) |
| 0.0 | 0.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0031331 | positive regulation of cellular catabolic process(GO:0031331) |
| 0.0 | 0.4 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.7 | GO:0042244 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.0 | 0.1 | GO:0002376 | immune effector process(GO:0002252) immune system process(GO:0002376) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.0 | 0.8 | GO:0016573 | internal protein amino acid acetylation(GO:0006475) histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) peptidyl-lysine acetylation(GO:0018394) |
| 0.0 | 0.2 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal metabolic process(GO:0009438) methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.0 | 0.1 | GO:0046503 | glycerophospholipid catabolic process(GO:0046475) glycerolipid catabolic process(GO:0046503) |
| 0.0 | 0.6 | GO:0000725 | recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.6 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.1 | GO:0019692 | deoxyribose phosphate metabolic process(GO:0019692) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.0 | 0.2 | GO:0007533 | mating type switching(GO:0007533) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.0 | 0.0 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.0 | 0.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) proton-transporting ATP synthase complex assembly(GO:0043461) |
| 0.0 | 0.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.5 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.0 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 | 0.2 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.1 | GO:0010529 | negative regulation of transposition, RNA-mediated(GO:0010526) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.4 | GO:0044843 | G1/S transition of mitotic cell cycle(GO:0000082) cell cycle G1/S phase transition(GO:0044843) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.0 | 0.2 | GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress(GO:0036003) |
| 0.0 | 0.2 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0043633 | polyadenylation-dependent RNA catabolic process(GO:0043633) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.0 | 0.0 | GO:0006784 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.2 | 0.9 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.1 | 0.9 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.3 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0000133 | polarisome(GO:0000133) |
| 0.1 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.2 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.1 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.2 | GO:0033551 | monopolin complex(GO:0033551) |
| 0.0 | 0.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.6 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.8 | GO:0005628 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.0 | 0.2 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) nucleoplasmic THO complex(GO:0000446) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase complex(GO:0000836) Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.0 | 0.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.0 | 1.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.5 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0031160 | ascospore wall(GO:0005619) spore wall(GO:0031160) |
| 0.0 | 0.1 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.0 | 0.1 | GO:0072380 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.0 | 0.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.1 | GO:0070211 | Snt2C complex(GO:0070211) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0044697 | HICS complex(GO:0044697) |
| 0.0 | 0.1 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.0 | 0.1 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) SAGA-type complex(GO:0070461) |
| 0.0 | 0.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0005825 | half bridge of spindle pole body(GO:0005825) |
| 0.0 | 1.3 | GO:0042995 | mating projection(GO:0005937) cell projection(GO:0042995) |
| 0.0 | 0.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.0 | GO:0033255 | SAS acetyltransferase complex(GO:0033255) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.0 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.0 | GO:0070274 | RES complex(GO:0070274) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005186 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.3 | 0.8 | GO:0005536 | glucose binding(GO:0005536) |
| 0.2 | 0.7 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.2 | 0.6 | GO:0008902 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.2 | 0.6 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.2 | 0.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.2 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.4 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.1 | 0.5 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.1 | 0.5 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.4 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 0.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.3 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.1 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.6 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.2 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.2 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.3 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 0.1 | GO:0019902 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.1 | 0.3 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.3 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 1.1 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.1 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 6.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0003916 | DNA topoisomerase activity(GO:0003916) |
| 0.0 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.3 | GO:0001031 | RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) transcription factor activity, RNA polymerase III promoter sequence-specific binding, TFIIIB recruiting(GO:0001004) transcription factor activity, RNA polymerase III type 1 promoter sequence-specific binding, TFIIIB recruiting(GO:0001005) transcription factor activity, RNA polymerase III type 2 promoter sequence-specific binding, TFIIIB recruiting(GO:0001008) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 3.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.0 | 0.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.0 | 0.1 | GO:0004375 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 1.4 | GO:0000988 | transcription factor activity, protein binding(GO:0000988) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) flap endonuclease activity(GO:0048256) |
| 0.0 | 0.0 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.0 | 0.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.0 | 0.1 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.0 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.1 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.0 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.7 | GO:0005543 | phospholipid binding(GO:0005543) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 23.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.1 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME RNA POL II TRANSCRIPTION | Genes involved in RNA Polymerase II Transcription |
| 0.0 | 23.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |