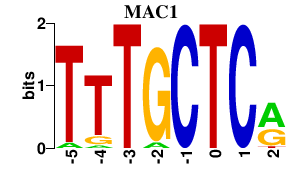
Results for MAC1
Z-value: 0.82
Transcription factors associated with MAC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAC1
|
S000004623 | Copper-sensing transcription factor |
Activity-expression correlation:
Activity profile of MAC1 motif
Sorted Z-values of MAC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YJL089W | 3.76 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YPR124W | 3.40 |
CTR1
|
High-affinity copper transporter of the plasma membrane, mediates nearly all copper uptake under low copper conditions; transcriptionally induced at low copper levels and degraded at high copper levels |
|
| YPR123C | 3.29 |
Hypothetical protein |
||
| YKL148C | 2.44 |
SDH1
|
Flavoprotein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YLR214W | 2.03 |
FRE1
|
Ferric reductase and cupric reductase, reduces siderophore-bound iron and oxidized copper prior to uptake by transporters; expression induced by low copper and iron levels |
|
| YIL162W | 1.90 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YMR271C | 1.77 |
URA10
|
Minor orotate phosphoribosyltransferase (OPRTase) isozyme that catalyzes the fifth enzymatic step in the de novo biosynthesis of pyrimidines, converting orotate into orotidine-5'-phosphate; major OPRTase encoded by URA5 |
|
| YKL147C | 1.76 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene AVT3 |
||
| YKR097W | 1.74 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YKL146W | 1.70 |
AVT3
|
Vacuolar transporter, exports large neutral amino acids from the vacuole; member of a family of seven S. cerevisiae genes (AVT1-7) related to vesicular GABA-glycine transporters |
|
| YAR053W | 1.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAL060W | 1.61 |
BDH1
|
NAD-dependent (R,R)-butanediol dehydrogenase, catalyzes oxidation of (R,R)-2,3-butanediol to (3R)-acetoin, oxidation of meso-butanediol to (3S)-acetoin, and reduction of acetoin; enhances use of 2,3-butanediol as an aerobic carbon source |
|
| YPL092W | 1.54 |
SSU1
|
Plasma membrane sulfite pump involved in sulfite metabolism and required for efficient sulfite efflux; major facilitator superfamily protein |
|
| YLR377C | 1.53 |
FBP1
|
Fructose-1,6-bisphosphatase, key regulatory enzyme in the gluconeogenesis pathway, required for glucose metabolism |
|
| YDR536W | 1.37 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YDL210W | 1.35 |
UGA4
|
Permease that serves as a gamma-aminobutyrate (GABA) transport protein involved in the utilization of GABA as a nitrogen source; catalyzes the transport of putrescine and delta-aminolevulinic acid (ALA); localized to the vacuolar membrane |
|
| YAR060C | 1.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YER065C | 1.29 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YFR053C | 1.28 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YCL001W-B | 1.24 |
Putative protein of unknown function; identified by homology |
||
| YIL033C | 1.20 |
BCY1
|
Regulatory subunit of the cyclic AMP-dependent protein kinase (PKA), a component of a signaling pathway that controls a variety of cellular processes, including metabolism, cell cycle, stress response, stationary phase, and sporulation |
|
| YMR090W | 1.16 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YCR005C | 1.12 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YPR054W | 1.09 |
SMK1
|
Middle sporulation-specific mitogen-activated protein kinase (MAPK) required for production of the outer spore wall layers; negatively regulates activity of the glucan synthase subunit Gsc2p |
|
| YDL182W | 1.06 |
LYS20
|
Homocitrate synthase isozyme, catalyzes the condensation of acetyl-CoA and alpha-ketoglutarate to form homocitrate, which is the first step in the lysine biosynthesis pathway; highly similar to the other isozyme, Lys21p |
|
| YGL255W | 1.05 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YOR343C | 1.02 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YGL256W | 1.00 |
ADH4
|
Alcohol dehydrogenase isoenzyme type IV, dimeric enzyme demonstrated to be zinc-dependent despite sequence similarity to iron-activated alcohol dehydrogenases; transcription is induced in response to zinc deficiency |
|
| YDR540C | 1.00 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YOR100C | 1.00 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YDR032C | 0.98 |
PST2
|
Protein with similarity to members of a family of flavodoxin-like proteins; induced by oxidative stress in a Yap1p dependent manner; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YMR297W | 0.98 |
PRC1
|
Vacuolar carboxypeptidase Y (proteinase C), broad-specificity C-terminal exopeptidase involved in non-specific protein degradation in the vacuole; member of the serine carboxypeptidase family |
|
| YJR115W | 0.94 |
Putative protein of unknown function |
||
| YLR213C | 0.94 |
CRR1
|
Putative glycoside hydrolase of the spore wall envelope; required for normal spore wall assembly, possibly for cross-linking between the glucan and chitosan layers; expressed during sporulation |
|
| YMR107W | 0.92 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YMR081C | 0.92 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YDL180W | 0.92 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YFL019C | 0.91 |
Dubious open reading frame unlikely to encode a protein; YFL019C is not an essential gene |
||
| YLR152C | 0.91 |
Putative protein of unknown function; YLR152C is not an essential gene |
||
| YFR047C | 0.90 |
BNA6
|
Quinolinate phosphoribosyl transferase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p |
|
| YBR069C | 0.89 |
TAT1
|
Amino acid transport protein for valine, leucine, isoleucine, and tyrosine, low-affinity tryptophan and histidine transporter; overexpression confers FK506 and FTY720 resistance |
|
| YPR001W | 0.89 |
CIT3
|
Dual specificity mitochondrial citrate and methylcitrate synthase; catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate and that of propionyl-CoA and oxaloacetate to form 2-methylcitrate |
|
| YFR017C | 0.88 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YFR017C is not an essential gene |
||
| YHR212C | 0.86 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR327C | 0.85 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YLR258W | 0.82 |
GSY2
|
Glycogen synthase, similar to Gsy1p; expression induced by glucose limitation, nitrogen starvation, heat shock, and stationary phase; activity regulated by cAMP-dependent, Snf1p and Pho85p kinases as well as by the Gac1p-Glc7p phosphatase |
|
| YGR191W | 0.81 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YPL229W | 0.80 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YPL229W is not an essential gene |
||
| YLR122C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YKL217W | 0.80 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YOR152C | 0.79 |
Putative protein of unknown function; has no similarity to any known protein; YOR152C is not an essential gene |
||
| YFL051C | 0.77 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YGR190C | 0.77 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YPR191W | 0.75 |
QCR2
|
Subunit 2 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; phosphorylated; transcription is regulated by Hap1p, Hap2p/Hap3p, and heme |
|
| YPL274W | 0.75 |
SAM3
|
High-affinity S-adenosylmethionine permease, required for utilization of S-adenosylmethionine as a sulfur source; has similarity to S-methylmethionine permease Mmp1p |
|
| YNL160W | 0.74 |
YGP1
|
Cell wall-related secretory glycoprotein; induced by nutrient deprivation-associated growth arrest and upon entry into stationary phase; may be involved in adaptation prior to stationary phase entry; has similarity to Sps100p |
|
| YPL171C | 0.73 |
OYE3
|
Widely conserved NADPH oxidoreductase containing flavin mononucleotide (FMN), homologous to Oye2p with slight differences in ligand binding and catalytic properties; may be involved in sterol metabolism |
|
| YER066W | 0.73 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YNR014W | 0.73 |
Putative protein of unknown function; expression is cell-cycle regulated, Azf1p-dependent, and heat-inducible |
||
| YNL194C | 0.73 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YAR047C | 0.72 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR319C | 0.72 |
FET4
|
Low-affinity Fe(II) transporter of the plasma membrane |
|
| YCL005W | 0.72 |
LDB16
|
Protein of unknown function; null mutants have decreased net negative cell surface charge; GFP-fusion protein expression is induced in response to the DNA-damaging agent MMS; native protein is detected in purified mitochondria |
|
| YLR123C | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YBR209W | 0.71 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR209W is not an essential gene |
||
| YFR007W | 0.70 |
YFH7
|
Putative kinase with similarity to the phosphoribulokinase/uridine kinase/bacterial pantothenate kinase (PRK/URK/PANK) subfamily of P-loop kinases; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YER076C | 0.70 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization |
||
| YCL048W-A | 0.70 |
Putative protein of unknown function |
||
| YJL062W-A | 0.70 |
Putative protein of unknown function, identified based on comparison to related yeast species; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YFL024C | 0.69 |
EPL1
|
Component of NuA4, which is an essential histone H4/H2A acetyltransferase complex; homologous to Drosophila Enhancer of Polycomb |
|
| YBR145W | 0.69 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YNL086W | 0.69 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to endosomes |
||
| YPL135W | 0.68 |
ISU1
|
Conserved protein of the mitochondrial matrix, performs a scaffolding function during assembly of iron-sulfur clusters, interacts physically and functionally with yeast frataxin (Yfh1p); isu1 isu2 double mutant is inviable |
|
| YMR089C | 0.68 |
YTA12
|
Component, with Afg3p, of the mitochondrial inner membrane m-AAA protease that mediates degradation of misfolded or unassembled proteins and is also required for correct assembly of mitochondrial enzyme complexes |
|
| YAL061W | 0.67 |
BDH2
|
Putative medium-chain alcohol dehydrogenase with similarity to BDH1; transcription induced by constitutively active PDR1 and PDR3; BDH2 is an essential gene |
|
| YCL038C | 0.67 |
ATG22
|
Vacuolar integral membrane protein required for efflux of amino acids during autophagic body breakdown in the vacuole; null mutation causes a gradual loss of viability during starvation |
|
| YPR196W | 0.67 |
Putative maltose activator |
||
| YLR203C | 0.67 |
MSS51
|
Nuclear encoded protein required for translation of COX1 mRNA; binds to Cox1 protein |
|
| YMR008C | 0.67 |
PLB1
|
Phospholipase B (lysophospholipase) involved in lipid metabolism, required for deacylation of phosphatidylcholine and phosphatidylethanolamine but not phosphatidylinositol |
|
| YER039C-A | 0.66 |
Putative protein of unknown function; YER039C-A is not an essential gene |
||
| YOR316C | 0.66 |
COT1
|
Vacuolar transporter that mediates zinc transport into the vacuole; overexpression confers resistance to cobalt and rhodium |
|
| YHR048W | 0.66 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YLR279W | 0.66 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL082W | 0.63 |
ATG19
|
Receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway; recognizes cargo proteins aminopeptidase I (Lap4p) and alpha-mannosidase (Ams1p) and delivers them to the preautophagosomal structure for packaging into Cvt vesicles |
|
| YNL046W | 0.62 |
Putative protein of unknown function; expression depends on Swi5p; GFP-fusion protein localizes to the endoplasmic reticulum; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YLR295C | 0.62 |
ATP14
|
Subunit h of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YDR411C | 0.62 |
DFM1
|
ER localized derlin-like family member involved in ER stress and homeostasis; not involved in ERAD or substrate retrotranslocation; interacts with CDC48; contains four transmembrane domains and two SHP boxes |
|
| YIL113W | 0.61 |
SDP1
|
Stress-inducible dual-specificity MAP kinase phosphatase, negatively regulates Slt2p MAP kinase by direct dephosphorylation, diffuse localization under normal conditions shifts to punctate localization after heat shock |
|
| YOR049C | 0.61 |
RSB1
|
Suppressor of sphingoid long chain base (LCB) sensitivity of an LCB-lyase mutation; putative integral membrane transporter or flippase that may transport LCBs from the cytoplasmic side toward the extracytoplasmic side of the membrane |
|
| YNL134C | 0.59 |
Putative protein of unknown function with similarity to dehydrogenases from other model organisms; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and nucleus and is induced by the DNA-damaging agent MMS |
||
| YLR259C | 0.59 |
HSP60
|
Tetradecameric mitochondrial chaperonin required for ATP-dependent folding of precursor polypeptides and complex assembly; prevents aggregation and mediates protein refolding after heat shock; role in mtDNA transmission; phosphorylated |
|
| YPL006W | 0.59 |
NCR1
|
Vacuolar membrane protein that transits through the biosynthetic vacuolar protein sorting pathway, involved in sphingolipid metabolism; glycoprotein and functional orthologue of human Niemann Pick C1 (NPC1) protein |
|
| YDR453C | 0.59 |
TSA2
|
Stress inducible cytoplasmic thioredoxin peroxidase; cooperates with Tsa1p in the removal of reactive oxygen, nitrogen and sulfur species using thioredoxin as hydrogen donor; deletion enhances the mutator phenotype of tsa1 mutants |
|
| YGL104C | 0.59 |
VPS73
|
Mitochondrial protein; mutation affects vacuolar protein sorting; putative transporter; member of the sugar porter family |
|
| YDR476C | 0.58 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YDR476C is not an essential gene |
||
| YMR013C | 0.58 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YGR174C | 0.58 |
CBP4
|
Mitochondrial protein required for assembly of ubiquinol cytochrome-c reductase complex (cytochrome bc1 complex); interacts with Cbp3p and function is partially redundant with that of Cbp3p |
|
| YGL062W | 0.58 |
PYC1
|
Pyruvate carboxylase isoform, cytoplasmic enzyme that converts pyruvate to oxaloacetate; highly similar to isoform Pyc2p but differentially regulated; mutations in the human homolog are associated with lactic acidosis |
|
| YAR050W | 0.58 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YGL040C | 0.58 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YBR035C | 0.58 |
PDX3
|
Pyridoxine (pyridoxamine) phosphate oxidase, has homologs in E. coli and Myxococcus xanthus; transcription is under the general control of nitrogen metabolism |
|
| YCL054W | 0.57 |
SPB1
|
AdoMet-dependent methyltransferase involved in rRNA processing and 60S ribosomal subunit maturation; methylates G2922 in the tRNA docking site of the large subunit rRNA and in the absence of snR52, U2921; suppressor of PAB1 mutants |
|
| YDR178W | 0.57 |
SDH4
|
Membrane anchor subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone |
|
| YDR342C | 0.57 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YOR192C | 0.56 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YNL014W | 0.56 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YNL200C | 0.56 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YBL015W | 0.54 |
ACH1
|
Acetyl-coA hydrolase, primarily localized to mitochondria; phosphorylated; required for acetate utilization and for diploid pseudohyphal growth |
|
| YBR053C | 0.54 |
Putative protein of unknown function; induced by cell wall perturbation |
||
| YIL060W | 0.54 |
Putative protein of unknown function; mutant accumulates less glycogen than does wild type; YIL060W is not an essential gene |
||
| YHR211W | 0.54 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YER038W-A | 0.53 |
FMP49
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 99% of ORF overlaps the verified gene HVG1; protein product detected in mitochondria |
|
| YHR116W | 0.53 |
COX23
|
Mitochondrial intermembrane space protein that functions in mitochondrial copper homeostasis, essential for functional cytochrome oxidase expression; homologous to Cox17p |
|
| YER076W-A | 0.53 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER076C |
||
| YNL144C | 0.53 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YOR120W | 0.53 |
GCY1
|
Putative NADP(+) coupled glycerol dehydrogenase, proposed to be involved in an alternative pathway for glycerol catabolism; member of the aldo-keto reductase (AKR) family |
|
| YDR516C | 0.53 |
EMI2
|
Non-essential protein of unknown function required for transcriptional induction of the early meiotic-specific transcription factor IME1; required for sporulation; expression is regulated by glucose-repression transcription factors Mig1/2p |
|
| YOR002W | 0.52 |
ALG6
|
Alpha 1,3 glucosyltransferase, involved in transfer of oligosaccharides from dolichyl pyrophosphate to asparagine residues of proteins during N-linked protein glycosylation; mutations in human ortholog are associated with disease |
|
| YIL087C | 0.52 |
LRC2
|
Putative protein of unknown function; protein is detected in purified mitochondria in high-throughput studies; null mutant displays decreased mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YOL084W | 0.51 |
PHM7
|
Protein of unknown function, expression is regulated by phosphate levels; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery and vacuole |
|
| YPR093C | 0.51 |
ASR1
|
Protein involved in a putative alcohol-responsive signaling pathway; accumulates in the nucleus under alcohol stress; contains a Ring/PHD finger domain |
|
| YIL121W | 0.51 |
QDR2
|
Multidrug transporter of the major facilitator superfamily, required for resistance to quinidine, barban, cisplatin, and bleomycin; may have a role in potassium uptake |
|
| YBL016W | 0.51 |
FUS3
|
Mitogen-activated serine/threonine protein kinase involved in mating; phosphoactivated by Ste7p; substrates include Ste12p, Far1p, Bni1p, Sst2p; inhibits invasive growth during mating by phosphorylating Tec1p, promoting its degradation |
|
| YPR127W | 0.51 |
Putative protein of unknown function; expression is activated by transcription factor YRM1/YOR172W; green fluorescent protein (GFP)-fusion protein localizes to both the cytoplasm and the nucleus |
||
| YPR013C | 0.51 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YDL183C | 0.51 |
Putative protein of unknown function; YDL183C is not an essential gene |
||
| YGR106C | 0.49 |
VOA1
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuolar memebrane |
|
| YEL011W | 0.48 |
GLC3
|
Glycogen branching enzyme, involved in glycogen accumulation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YKL093W | 0.48 |
MBR1
|
Protein involved in mitochondrial functions and stress response; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants |
|
| YHR055C | 0.48 |
CUP1-2
|
Metallothionein, binds copper and mediates resistance to high concentrations of copper and cadmium; locus is variably amplified in different strains, with two copies, CUP1-1 and CUP1-2, in the genomic sequence reference strain S288C |
|
| YMR133W | 0.48 |
REC114
|
Protein involved in early stages of meiotic recombination; possibly involved in the coordination of recombination and meiotic division; mutations lead to premature initiation of the first meiotic division |
|
| YMR320W | 0.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKR102W | 0.47 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YOR065W | 0.47 |
CYT1
|
Cytochrome c1, component of the mitochondrial respiratory chain; expression is regulated by the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex |
|
| YJR048W | 0.47 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YGL143C | 0.46 |
MRF1
|
Mitochondrial translation release factor, involved in stop codon recognition and hydrolysis of the peptidyl-tRNA bond during mitochondrial translation; lack of MRF1 causes mitochondrial genome instability |
|
| YPL117C | 0.46 |
IDI1
|
Isopentenyl diphosphate:dimethylallyl diphosphate isomerase (IPP isomerase), catalyzes an essential activation step in the isoprenoid biosynthetic pathway; required for viability |
|
| YBR140C | 0.46 |
IRA1
|
GTPase-activating protein that negatively regulates RAS by converting it from the GTP- to the GDP-bound inactive form, required for reducing cAMP levels under nutrient limiting conditions, mediates membrane association of adenylate cyclase |
|
| YIL059C | 0.46 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YIL060W |
||
| YGR239C | 0.45 |
PEX21
|
Peroxin required for targeting of peroxisomal matrix proteins containing PTS2; interacts with Pex7p; partially redundant with Pex18p |
|
| YLR125W | 0.45 |
Putative protein of unknown function; mutant has decreased Ty3 transposition; YLR125W is not an essential gene |
||
| YLR004C | 0.45 |
THI73
|
Putative plasma membrane permease proposed to be involved in carboxylic acid uptake and repressed by thiamine; substrate of Dbf2p/Mob1p kinase; transcription is altered if mitochondrial dysfunction occurs |
|
| YDR036C | 0.45 |
EHD3
|
3-hydroxyisobutyryl-CoA hydrolase, member of a family of enoyl-CoA hydratase/isomerases; non-tagged protein is detected in highly purified mitochondria in high-throughput studies; phosphorylated; mutation affects fluid-phase endocytosis |
|
| YGL184C | 0.45 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YCR021C | 0.45 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YIL032C | 0.45 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR027C | 0.44 |
Putative protein of unknown function |
||
| YEL024W | 0.44 |
RIP1
|
Ubiquinol-cytochrome-c reductase, a Rieske iron-sulfur protein of the mitochondrial cytochrome bc1 complex; transfers electrons from ubiquinol to cytochrome c1 during respiration |
|
| YLR356W | 0.43 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YDL072C | 0.43 |
YET3
|
Protein of unknown function; YET3 null mutant decreases the level of secreted invertase; homolog of human BAP31 protein |
|
| YHR139C | 0.43 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YBL102W | 0.43 |
SFT2
|
Non-essential tetra-spanning membrane protein found mostly in the late Golgi, can suppress some sed5 alleles; may be part of the transport machinery, but precise function is unknown; similar to mammalian syntaxin 5 |
|
| YML123C | 0.42 |
PHO84
|
High-affinity inorganic phosphate (Pi) transporter and low-affinity manganese transporter; regulated by Pho4p and Spt7p; mutation confers resistance to arsenate; exit from the ER during maturation requires Pho86p |
|
| YIL058W | 0.42 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER053C | 0.42 |
PIC2
|
Mitochondrial phosphate carrier, imports inorganic phosphate into mitochondria; functionally redundant with Mir1p but less abundant than Mir1p under normal conditions; expression is induced at high temperature |
|
| YOL085W-A | 0.42 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YOL085C |
||
| YNR050C | 0.41 |
LYS9
|
Saccharopine dehydrogenase (NADP+, L-glutamate-forming); catalyzes the formation of saccharopine from alpha-aminoadipate 6-semialdehyde, which is the seventh step in lysine biosynthesis pathway |
|
| YHR053C | 0.41 |
CUP1-1
|
Metallothionein, binds copper and mediates resistance to high concentrations of copper and cadmium; locus is variably amplified in different strains, with two copies, CUP1-1 and CUP1-2, in the genomic sequence reference strain S288C |
|
| YLR030W | 0.41 |
Putative protein of unknown function |
||
| YPL048W | 0.41 |
CAM1
|
Nuclear protein required for transcription of MXR1; binds the MXR1 promoter in the presence of other nuclear factors; binds calcium and phospholipids; has similarity to translational cofactor EF-1 gamma |
|
| YKL038W | 0.40 |
RGT1
|
Glucose-responsive transcription factor that regulates expression of several glucose transporter (HXT) genes in response to glucose; binds to promoters and acts both as a transcriptional activator and repressor |
|
| YMR135C | 0.40 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YOL154W | 0.40 |
ZPS1
|
Putative GPI-anchored protein; transcription is induced under low-zinc conditions, as mediated by the Zap1p transcription factor, and at alkaline pH |
|
| YAR068W | 0.40 |
Fungal-specific protein of unknown function; induced in respiratory-deficient cells |
||
| YJL172W | 0.40 |
CPS1
|
Vacuolar carboxypeptidase yscS; expression is induced under low-nitrogen conditions |
|
| YER133W-A | 0.40 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps uncharacterized gene YER134C. |
||
| YHR202W | 0.40 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole, while HA-tagged protein is found in the soluble fraction, suggesting cytoplasmic localization |
||
| YJL093C | 0.39 |
TOK1
|
Outward-rectifier potassium channel of the plasma membrane with two pore domains in tandem, each of which forms a functional channel permeable to potassium; carboxy tail functions to prevent inner gate closures; target of K1 toxin |
|
| YOR149C | 0.39 |
SMP3
|
Alpha 1,2-mannosyltransferase involved in glycosyl phosphatidyl inositol (GPI) biosynthesis; required for addition of the fourth, side branching mannose to the GPI core structure |
|
| YLR228C | 0.39 |
ECM22
|
Sterol regulatory element binding protein, regulates transcription of the sterol biosynthetic genes ERG2 and ERG3; member of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors; homologous to Upc2p |
|
| YLR374C | 0.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF STP3/YLR375W |
||
| YBL049W | 0.39 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YDR249C | 0.38 |
Putative protein of unknown function |
||
| YJR095W | 0.38 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YHR054C | 0.38 |
Putative protein of unknown function |
||
| YLR375W | 0.38 |
STP3
|
Zinc-finger protein of unknown function, possibly involved in pre-tRNA splicing and in uptake of branched-chain amino acids |
|
| YDR530C | 0.38 |
APA2
|
Diadenosine 5',5''-P1,P4-tetraphosphate phosphorylase II (AP4A phosphorylase), involved in catabolism of bis(5'-nucleosidyl) tetraphosphates; has similarity to Apa1p |
|
| YNL052W | 0.38 |
COX5A
|
Subunit Va of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during aerobic growth while its isoform Vb (Cox5Bp) is expressed during anaerobic growth |
|
| YMR042W | 0.37 |
ARG80
|
Transcription factor involved in regulation of arginine-responsive genes; acts with Arg81p and Arg82p |
|
| YGR022C | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YMR206W | 0.37 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YLR395C | 0.37 |
COX8
|
Subunit VIII of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YFL020C | 0.37 |
PAU5
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YNL288W | 0.37 |
CAF40
|
Evolutionarily conserved subunit of the CCR4-NOT complex involved in controlling mRNA initiation, elongation and degradation; binds Cdc39p |
|
| YNL179C | 0.36 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YBR050C | 0.36 |
REG2
|
Regulatory subunit of the Glc7p type-1 protein phosphatase; involved with Reg1p, Glc7p, and Snf1p in regulation of glucose-repressible genes, also involved in glucose-induced proteolysis of maltose permease |
|
| YDR298C | 0.36 |
ATP5
|
Subunit 5 of the stator stalk of mitochondrial F1F0 ATP synthase, which is an evolutionarily conserved enzyme complex required for ATP synthesis; homologous to bovine subunit OSCP (oligomycin sensitivity-conferring protein); phosphorylated |
|
| YBL017C | 0.36 |
PEP1
|
Type I transmembrane sorting receptor for multiple vacuolar hydrolases; cycles between the late-Golgi and prevacuolar endosome-like compartments |
|
| YGR102C | 0.36 |
Putative protein of unknown function; transposon insertion mutant is salt sensitive and deletion mutant has growth defects; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YGR110W | 0.36 |
Putative protein of unknown function; transcription is increased in response to genotoxic stress; plays a role in restricting Ty1 transposition |
||
| YPL207W | 0.35 |
TYW1
|
Protein required for the synthesis of wybutosine, a modified guanosine found at the 3'-position adjacent to the anticodon of phenylalanine tRNA which supports reading frame maintenance by stabilizing codon-anticodon interactions |
|
| YNL180C | 0.35 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YLR296W | 0.35 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL060C | 0.35 |
MAM3
|
Protein required for normal mitochondrial morphology, has similarity to hemolysins |
|
| YPR160W | 0.34 |
GPH1
|
Non-essential glycogen phosphorylase required for the mobilization of glycogen, activity is regulated by cyclic AMP-mediated phosphorylation, expression is regulated by stress-response elements and by the HOG MAP kinase pathway |
|
| YGR149W | 0.34 |
Putative protein of unknown function; predicted to be an integal membrane protein |
||
| YER033C | 0.34 |
ZRG8
|
Protein of unknown function; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; GFP-fusion protein is localized to the cytoplasm; transcription induced under conditions of zinc deficiency |
|
| YBR220C | 0.34 |
Putative protein of unknown function; YBR220C is not an essential gene |
||
| YJL217W | 0.34 |
Cytoplasmic protein of unknown function; expression induced by calcium shortage and via the copper sensing transciption factor Mac1p during conditons of copper deficiency; mRNA is cell cycle regulated, peaking in G1 phase |
||
| YJL088W | 0.34 |
ARG3
|
Ornithine carbamoyltransferase (carbamoylphosphate:L-ornithine carbamoyltransferase), catalyzes the sixth step in the biosynthesis of the arginine precursor ornithine |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAC1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.8 | 6.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.6 | 3.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.5 | 2.0 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.5 | 2.4 | GO:0034487 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.4 | 1.3 | GO:0015755 | fructose transport(GO:0015755) |
| 0.4 | 1.1 | GO:0019626 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.4 | 1.1 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.3 | 1.3 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.3 | 2.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.8 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.3 | 1.5 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.3 | 1.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.3 | 0.8 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.3 | 1.1 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.3 | 1.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 0.8 | GO:0045117 | azole transport(GO:0045117) |
| 0.3 | 2.0 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.2 | 1.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 2.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.7 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
| 0.2 | 1.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.4 | GO:0046323 | glucose import(GO:0046323) |
| 0.2 | 1.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.2 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.2 | 0.4 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.2 | 1.9 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.2 | 0.7 | GO:0046352 | disaccharide catabolic process(GO:0046352) |
| 0.2 | 1.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 2.4 | GO:0072593 | reactive oxygen species metabolic process(GO:0072593) |
| 0.1 | 0.6 | GO:0032373 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) positive regulation of sterol transport(GO:0032373) positive regulation of transmembrane transport(GO:0034764) regulation of sterol import by regulation of transcription from RNA polymerase II promoter(GO:0035968) positive regulation of sterol import by positive regulation of transcription from RNA polymerase II promoter(GO:0035969) regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.1 | 0.6 | GO:1903137 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.1 | 0.8 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.1 | 0.4 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.1 | 1.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 0.9 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 0.4 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 1.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.6 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 1.7 | GO:0046173 | polyol biosynthetic process(GO:0046173) |
| 0.1 | 0.3 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.1 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) regulation of cellular amine catabolic process(GO:0033241) |
| 0.1 | 0.6 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.8 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.4 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.6 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.1 | 0.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.5 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.1 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.8 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.1 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 2.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.4 | GO:0009209 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.1 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.3 | GO:0009450 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.4 | GO:0016114 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.3 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.1 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.1 | 1.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.1 | 0.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.5 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.1 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.5 | GO:0097034 | mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.6 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.1 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.8 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.1 | 0.5 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0032108 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.1 | 2.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.2 | GO:0015749 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.1 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.6 | GO:0006071 | glycerol metabolic process(GO:0006071) alditol metabolic process(GO:0019400) |
| 0.1 | 0.5 | GO:0009306 | protein secretion(GO:0009306) |
| 0.1 | 1.0 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.2 | GO:0006032 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.3 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.2 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:1903513 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.1 | 0.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.3 | GO:0006074 | (1->3)-beta-D-glucan metabolic process(GO:0006074) |
| 0.1 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.1 | GO:0015848 | spermidine transport(GO:0015848) |
| 0.1 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.1 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.0 | 1.1 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.2 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.2 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 1.5 | GO:0070726 | cell wall assembly(GO:0070726) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:2000001 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.3 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.2 | GO:0008299 | isoprenoid metabolic process(GO:0006720) isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) post-translational protein modification(GO:0043687) |
| 0.0 | 0.1 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 0.1 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.2 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.7 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.1 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.0 | 0.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0009187 | cyclic nucleotide metabolic process(GO:0009187) |
| 0.0 | 0.1 | GO:0000736 | double-strand break repair via single-strand annealing, removal of nonhomologous ends(GO:0000736) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 | 0.1 | GO:0055078 | cellular sodium ion homeostasis(GO:0006883) sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.2 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.0 | GO:0035384 | thioester biosynthetic process(GO:0035384) acyl-CoA biosynthetic process(GO:0071616) |
| 0.0 | 0.2 | GO:0030846 | termination of RNA polymerase II transcription, poly(A)-coupled(GO:0030846) |
| 0.0 | 0.1 | GO:0010821 | regulation of mitochondrion organization(GO:0010821) |
| 0.0 | 0.2 | GO:0051453 | regulation of cellular pH(GO:0030641) regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:0051274 | beta-glucan biosynthetic process(GO:0051274) |
| 0.0 | 0.5 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.0 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.5 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.0 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.0 | 0.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.0 | GO:0010978 | gene silencing involved in chronological cell aging(GO:0010978) gene silencing by RNA(GO:0031047) |
| 0.0 | 0.1 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) calcium ion homeostasis(GO:0055074) |
| 0.0 | 0.0 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.0 | GO:0036095 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.0 | 0.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0043112 | receptor recycling(GO:0001881) protein import into peroxisome matrix, receptor recycling(GO:0016562) receptor metabolic process(GO:0043112) |
| 0.0 | 0.0 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.0 | 0.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.1 | GO:0046834 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0065005 | lipid tube assembly(GO:0060988) protein-lipid complex assembly(GO:0065005) protein-lipid complex subunit organization(GO:0071825) |
| 0.0 | 0.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.0 | GO:0045991 | carbon catabolite activation of transcription(GO:0045991) |
| 0.0 | 0.0 | GO:1903727 | positive regulation of phospholipid biosynthetic process(GO:0071073) positive regulation of phospholipid metabolic process(GO:1903727) positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.0 | 0.1 | GO:0019217 | regulation of fatty acid metabolic process(GO:0019217) |
| 0.0 | 0.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.3 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 | 0.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 2.8 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.3 | 0.7 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.3 | 0.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 0.7 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.2 | 0.6 | GO:0045269 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.2 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.4 | GO:0044284 | mitochondrial crista junction(GO:0044284) |
| 0.1 | 3.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.0 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.1 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.6 | GO:0000837 | Doa10p ubiquitin ligase complex(GO:0000837) |
| 0.1 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.4 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.1 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.1 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0033177 | proton-transporting two-sector ATPase complex, proton-transporting domain(GO:0033177) |
| 0.1 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.1 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 2.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.2 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.1 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.2 | GO:0033100 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 1.3 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.0 | 0.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0030139 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0048188 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 3.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.0 | GO:0031160 | spore wall(GO:0031160) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 20.8 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.1 | GO:0044455 | mitochondrial membrane part(GO:0044455) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0071561 | nucleus-vacuole junction(GO:0071561) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0070823 | HDA1 complex(GO:0070823) |
| 0.0 | 0.1 | GO:0000500 | RNA polymerase I upstream activating factor complex(GO:0000500) |
| 0.0 | 0.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.8 | 2.3 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.8 | 3.8 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.7 | 2.1 | GO:0004575 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.6 | 3.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.6 | 1.7 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) L-glutamine transmembrane transporter activity(GO:0015186) L-isoleucine transmembrane transporter activity(GO:0015188) branched-chain amino acid transmembrane transporter activity(GO:0015658) |
| 0.5 | 1.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.5 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 1.8 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 1.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 1.9 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.4 | 0.7 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.3 | 1.0 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.3 | 1.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.3 | 0.9 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.3 | 2.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.3 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 2.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 1.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 0.7 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.2 | 1.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 1.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 2.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 1.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.5 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.2 | 0.6 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.2 | 0.5 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 1.8 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.4 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.1 | 1.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 0.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.4 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.1 | 1.2 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0016406 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.4 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.5 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.8 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.4 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 0.4 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.1 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.1 | 0.1 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.1 | 0.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.4 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.1 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.3 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.1 | 0.2 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.4 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.1 | 0.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.2 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III transcription factor binding(GO:0001025) TFIIIB-type transcription factor activity(GO:0001026) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.2 | GO:0001128 | RNA polymerase II transcription coactivator activity involved in preinitiation complex assembly(GO:0001128) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0016898 | lactate dehydrogenase activity(GO:0004457) D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 2.5 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.0 | GO:0016878 | CoA-ligase activity(GO:0016405) acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.0 | GO:0022829 | porin activity(GO:0015288) wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 1.0 | GO:0061733 | histone acetyltransferase activity(GO:0004402) peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.9 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.5 | GO:0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors(GO:0016903) |
| 0.0 | 0.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0032934 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.0 | 0.3 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.0 | GO:0015085 | calcium ion transmembrane transporter activity(GO:0015085) |
| 0.0 | 0.2 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0097617 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.0 | 0.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.0 | 0.1 | GO:0001168 | transcription factor activity, RNA polymerase I upstream control element sequence-specific binding(GO:0001168) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0015037 | glutathione-disulfide reductase activity(GO:0004362) peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 25.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.1 | 0.4 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.1 | 0.1 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.1 | 0.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 25.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |