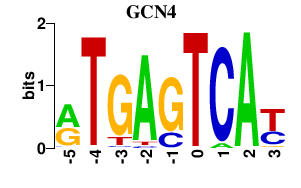
Results for GCN4
Z-value: 1.05
Transcription factors associated with GCN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GCN4
|
S000000735 | bZIP transcriptional activator of amino acid biosynthetic genes |
Activity-expression correlation:
Activity profile of GCN4 motif
Sorted Z-values of GCN4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YLR307C-A | 9.02 |
Putative protein of unknown function |
||
| YPL250C | 6.69 |
ICY2
|
Protein of unknown function; mobilized into polysomes upon a shift from a fermentable to nonfermentable carbon source; potential Cdc28p substrate |
|
| YAR053W | 6.13 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YMR096W | 5.61 |
SNZ1
|
Protein involved in vitamin B6 biosynthesis; member of a stationary phase-induced gene family; coregulated with SNO1; interacts with Sno1p and with Yhr198p, perhaps as a multiprotein complex containing other Snz and Sno proteins |
|
| YAR060C | 4.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL104C | 4.54 |
LEU4
|
Alpha-isopropylmalate synthase (2-isopropylmalate synthase); the main isozyme responsible for the first step in the leucine biosynthesis pathway |
|
| YHR212C | 4.03 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR116C | 3.96 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene TKL2 |
||
| YJL088W | 3.77 |
ARG3
|
Ornithine carbamoyltransferase (carbamoylphosphate:L-ornithine carbamoyltransferase), catalyzes the sixth step in the biosynthesis of the arginine precursor ornithine |
|
| YMR107W | 3.74 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YLR356W | 3.71 |
Putative protein of unknown function with similarity to SCM4; green fluorescent protein (GFP)-fusion protein localizes to mitochondria; YLR356W is not an essential gene |
||
| YPR036W-A | 3.66 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YMR095C | 3.66 |
SNO1
|
Protein of unconfirmed function, involved in pyridoxine metabolism; expression is induced during stationary phase; forms a putative glutamine amidotransferase complex with Snz1p, with Sno1p serving as the glutaminase |
|
| YBR145W | 3.56 |
ADH5
|
Alcohol dehydrogenase isoenzyme V; involved in ethanol production |
|
| YDL182W | 3.55 |
LYS20
|
Homocitrate synthase isozyme, catalyzes the condensation of acetyl-CoA and alpha-ketoglutarate to form homocitrate, which is the first step in the lysine biosynthesis pathway; highly similar to the other isozyme, Lys21p |
|
| YPL092W | 3.48 |
SSU1
|
Plasma membrane sulfite pump involved in sulfite metabolism and required for efficient sulfite efflux; major facilitator superfamily protein |
|
| YJR146W | 3.47 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YKL217W | 3.46 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YHR212W-A | 3.40 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YLR327C | 3.40 |
TMA10
|
Protein of unknown function that associates with ribosomes |
|
| YER068C-A | 3.38 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL162W | 3.21 |
SUC2
|
Invertase, sucrose hydrolyzing enzyme; a secreted, glycosylated form is regulated by glucose repression, and an intracellular, nonglycosylated enzyme is produced constitutively |
|
| YER069W | 3.16 |
ARG5,6
|
Protein that is processed in the mitochondrion to yield acetylglutamate kinase and N-acetyl-gamma-glutamyl-phosphate reductase, which catalyze the 2nd and 3rd steps in arginine biosynthesis; enzymes form a complex with Arg2p |
|
| YCL030C | 2.94 |
HIS4
|
Multifunctional enzyme containing phosphoribosyl-ATP pyrophosphatase, phosphoribosyl-AMP cyclohydrolase, and histidinol dehydrogenase activities; catalyzes the second, third, ninth and tenth steps in histidine biosynthesis |
|
| YHR071W | 2.86 |
PCL5
|
Cyclin, interacts with Pho85p cyclin-dependent kinase (Cdk), induced by Gcn4p at level of transcription, specifically required for Gcn4p degradation, may be sensor of cellular protein biosynthetic capacity |
|
| YOL058W | 2.82 |
ARG1
|
Arginosuccinate synthetase, catalyzes the formation of L-argininosuccinate from citrulline and L-aspartate in the arginine biosynthesis pathway; potential Cdc28p substrate |
|
| YPR184W | 2.74 |
GDB1
|
Glycogen debranching enzyme containing glucanotranferase and alpha-1,6-amyloglucosidase activities, required for glycogen degradation; phosphorylated in mitochondria |
|
| YBR147W | 2.67 |
RTC2
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays fluconazole resistance and suppresses cdc13-1 temperature sensitivity |
|
| YDR540C | 2.67 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YFR052C-A | 2.65 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR533C | 2.59 |
HSP31
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site |
|
| YNL005C | 2.57 |
MRP7
|
Mitochondrial ribosomal protein of the large subunit |
|
| YPL018W | 2.49 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YKL016C | 2.46 |
ATP7
|
Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YDL067C | 2.27 |
COX9
|
Subunit VIIa of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain |
|
| YDL183C | 2.24 |
Putative protein of unknown function; YDL183C is not an essential gene |
||
| YGL184C | 2.20 |
STR3
|
Cystathionine beta-lyase, converts cystathionine into homocysteine |
|
| YHL046W-A | 2.18 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR095W | 2.17 |
SFC1
|
Mitochondrial succinate-fumarate transporter, transports succinate into and fumarate out of the mitochondrion; required for ethanol and acetate utilization |
|
| YFL051C | 2.14 |
Putative protein of unknown function; YFL051C is not an essential gene |
||
| YER175C | 2.12 |
TMT1
|
Trans-aconitate methyltransferase, cytosolic enzyme that catalyzes the methyl esterification of 3-isopropylmalate, an intermediate of the leucine biosynthetic pathway, and trans-aconitate, which inhibits the citric acid cycle |
|
| YGR065C | 2.10 |
VHT1
|
High-affinity plasma membrane H+-biotin (vitamin H) symporter; mutation results in fatty acid auxotrophy; 12 transmembrane domain containing major facilitator subfamily member; mRNA levels negatively regulated by iron deprivation and biotin |
|
| YDL170W | 2.03 |
UGA3
|
Transcriptional activator necessary for gamma-aminobutyrate (GABA)-dependent induction of GABA genes (such as UGA1, UGA2, UGA4); zinc-finger transcription factor of the Zn(2)-Cys(6) binuclear cluster domain type; localized to the nucleus |
|
| YIR034C | 1.93 |
LYS1
|
Saccharopine dehydrogenase (NAD+, L-lysine-forming), catalyzes the conversion of saccharopine to L-lysine, which is the final step in the lysine biosynthesis pathway |
|
| YER024W | 1.91 |
YAT2
|
Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane |
|
| YKL103C | 1.84 |
LAP4
|
Vacuolar aminopeptidase, often used as a marker protein in studies of autophagy and cytosol to vacuole targeting (CVT) pathway |
|
| YHR048W | 1.81 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YHL047C | 1.79 |
ARN2
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to the siderophore triacetylfusarinine C |
|
| YNR069C | 1.76 |
BSC5
|
Protein of unknown function, ORF exhibits genomic organization compatible with a translational readthrough-dependent mode of expression |
|
| YHL036W | 1.75 |
MUP3
|
Low affinity methionine permease, similar to Mup1p |
|
| YCR005C | 1.74 |
CIT2
|
Citrate synthase, catalyzes the condensation of acetyl coenzyme A and oxaloacetate to form citrate, peroxisomal isozyme involved in glyoxylate cycle; expression is controlled by Rtg1p and Rtg2p transcription factors |
|
| YNL036W | 1.67 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YER065C | 1.62 |
ICL1
|
Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose |
|
| YOR100C | 1.62 |
CRC1
|
Mitochondrial inner membrane carnitine transporter, required for carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria during fatty acid beta-oxidation |
|
| YAL062W | 1.60 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YNL183C | 1.59 |
NPR1
|
Protein kinase that stabilizes several plasma membrane amino acid transporters by antagonizing their ubiquitin-mediated degradation |
|
| YOL118C | 1.58 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YHR092C | 1.57 |
HXT4
|
High-affinity glucose transporter of the major facilitator superfamily, expression is induced by low levels of glucose and repressed by high levels of glucose |
|
| YKL102C | 1.55 |
Dubious open reading frame unlikely to encode a functional protein; deletion confers sensitivity to citric acid; predicted protein would include a thiol-disulfide oxidoreductase active site |
||
| YMR081C | 1.54 |
ISF1
|
Serine-rich, hydrophilic protein with similarity to Mbr1p; overexpression suppresses growth defects of hap2, hap3, and hap4 mutants; expression is under glucose control; cotranscribed with NAM7 in a cyp1 mutant |
|
| YJR147W | 1.53 |
HMS2
|
Protein with similarity to heat shock transcription factors; overexpression suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YJL133C-A | 1.50 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YNR050C | 1.50 |
LYS9
|
Saccharopine dehydrogenase (NADP+, L-glutamate-forming); catalyzes the formation of saccharopine from alpha-aminoadipate 6-semialdehyde, which is the seventh step in lysine biosynthesis pathway |
|
| YNL125C | 1.50 |
ESBP6
|
Protein with similarity to monocarboxylate permeases, appears not to be involved in transport of monocarboxylates such as lactate, pyruvate or acetate across the plasma membrane |
|
| YFR029W | 1.49 |
PTR3
|
Component of the SPS plasma membrane amino acid sensor system (Ssy1p-Ptr3p-Ssy5p), which senses external amino acid concentration and transmits intracellular signals that result in regulation of expression of amino acid permease genes |
|
| YAR050W | 1.49 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YJL199C | 1.47 |
MBB1
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; protein detected in large-scale protein-protein interaction studies |
|
| YPL135W | 1.45 |
ISU1
|
Conserved protein of the mitochondrial matrix, performs a scaffolding function during assembly of iron-sulfur clusters, interacts physically and functionally with yeast frataxin (Yfh1p); isu1 isu2 double mutant is inviable |
|
| YOL140W | 1.44 |
ARG8
|
Acetylornithine aminotransferase, catalyzes the fourth step in the biosynthesis of the arginine precursor ornithine |
|
| YFR053C | 1.43 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YJR109C | 1.42 |
CPA2
|
Large subunit of carbamoyl phosphate synthetase, which catalyzes a step in the synthesis of citrulline, an arginine precursor |
|
| YLR152C | 1.40 |
Putative protein of unknown function; YLR152C is not an essential gene |
||
| YER175W-A | 1.39 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YPR010C-A | 1.39 |
Putative protein of unknown function; conserved among Saccharomyces sensu stricto species |
||
| YAR047C | 1.35 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR191W | 1.35 |
HIP1
|
High-affinity histidine permease, also involved in the transport of manganese ions |
|
| YOL119C | 1.31 |
MCH4
|
Protein with similarity to mammalian monocarboxylate permeases, which are involved in transport of monocarboxylic acids across the plasma membrane; mutant is not deficient in monocarboxylate transport |
|
| YDR258C | 1.31 |
HSP78
|
Oligomeric mitochondrial matrix chaperone that cooperates with Ssc1p in mitochondrial thermotolerance after heat shock; prevents the aggregation of misfolded matrix proteins; component of the mitochondrial proteolysis system |
|
| YBR218C | 1.29 |
PYC2
|
Pyruvate carboxylase isoform, cytoplasmic enzyme that converts pyruvate to oxaloacetate; highly similar to isoform Pyc1p but differentially regulated; mutations in the human homolog are associated with lactic acidosis |
|
| YBR250W | 1.25 |
SPO23
|
Protein of unknown function; associates with meiosis-specific protein Spo1p |
|
| YJR048W | 1.23 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YGL059W | 1.20 |
PKP2
|
Mitochondrial protein kinase that negatively regulates activity of the pyruvate dehydrogenase complex by phosphorylating the ser-133 residue of the Pda1p subunit; acts in concert with kinase Pkp1p and phosphatases Ptc5p and Ptc6p |
|
| YHL040C | 1.19 |
ARN1
|
Transporter, member of the ARN family of transporters that specifically recognize siderophore-iron chelates; responsible for uptake of iron bound to ferrirubin, ferrirhodin, and related siderophores |
|
| YDR010C | 1.18 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YIL047C | 1.15 |
SYG1
|
Plasma membrane protein of unknown function; truncation and overexpression suppresses lethality of G-alpha protein deficiency |
|
| YGR190C | 1.14 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene HIP1/YGR191W |
||
| YML076C | 1.13 |
WAR1
|
Homodimeric Zn2Cys6 zinc finger transcription factor; binds to a weak acid response element to induce transcription of PDR12 and FUN34, encoding an acid transporter and a putative ammonia transporter, respectively |
|
| YDR342C | 1.12 |
HXT7
|
High-affinity glucose transporter of the major facilitator superfamily, nearly identical to Hxt6p, expressed at high basal levels relative to other HXTs, expression repressed by high glucose levels |
|
| YMR114C | 1.10 |
Protein of unknown function; may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and cytoplasm; YMR114C is not an essential gene |
||
| YIL057C | 1.10 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YOL025W | 1.06 |
LAG2
|
Protein involved in determination of longevity; LAG2 gene is preferentially expressed in young cells; overexpression extends the mean and maximum life span of cells |
|
| YDR453C | 1.06 |
TSA2
|
Stress inducible cytoplasmic thioredoxin peroxidase; cooperates with Tsa1p in the removal of reactive oxygen, nitrogen and sulfur species using thioredoxin as hydrogen donor; deletion enhances the mutator phenotype of tsa1 mutants |
|
| YGR258C | 1.02 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YKR097W | 1.01 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YDL085W | 0.99 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YMR013C | 0.99 |
SEC59
|
Dolichol kinase, catalyzes the terminal step in dolichyl monophosphate (Dol-P) biosynthesis; required for viability and for normal rates of lipid intermediate synthesis and protein N-glycosylation |
|
| YNL037C | 0.99 |
IDH1
|
Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle |
|
| YCR023C | 0.99 |
Vacuolar membrane protein of unknown function; member of the multidrug resistance family; YCR023C is not an essential gene |
||
| YGL219C | 0.97 |
MDM34
|
Mitochondrial outer membrane protein, required for normal mitochondrial morphology and inheritance; localizes to dots on the mitochondrial surface near mtDNA nucleoids; interacts genetically with MDM31 and MDM32 |
|
| YER039C-A | 0.96 |
Putative protein of unknown function; YER039C-A is not an essential gene |
||
| YBR169C | 0.96 |
SSE2
|
Member of the heat shock protein 70 (HSP70) family; may be involved in protein folding; localized to the cytoplasm; highly homologous to the heat shock protein Sse1p |
|
| YJL185C | 0.95 |
Putative protein of unknown function; mRNA is weakly cell cycle regulated, peaking in G2 phase; YJL185C is a non-essential gene |
||
| YFL030W | 0.94 |
AGX1
|
Alanine:glyoxylate aminotransferase (AGT), catalyzes the synthesis of glycine from glyoxylate, which is one of three pathways for glycine biosynthesis in yeast; has similarity to mammalian and plant alanine:glyoxylate aminotransferases |
|
| YNL013C | 0.93 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF HEF3/YNL014W |
||
| YLL055W | 0.91 |
YCT1
|
High-affinity cysteine-specific transporter with similarity to the Dal5p family of transporters; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YCT1 is not an essential gene |
|
| YKL112W | 0.90 |
ABF1
|
DNA binding protein with possible chromatin-reorganizing activity involved in transcriptional activation, gene silencing, and DNA replication and repair |
|
| YJL213W | 0.90 |
Protein of unknown function that may interact with ribosomes; periodically expressed during the yeast metabolic cycle; phosphorylated in vitro by the mitotic exit network (MEN) kinase complex, Dbf2p/Mob1p |
||
| YLL029W | 0.90 |
FRA1
|
Protein involved in negative regulation of transcription of iron regulon; forms an iron independent complex with Fra2p, Grx3p, and Grx4p; cytosolic; mutant fails to repress transcription of iron regulon and is defective in spore formation |
|
| YJR122W | 0.89 |
IBA57
|
Mitochondrial matrix protein involved in the incorporation of iron-sulfur clusters into mitochondrial aconitase-type proteins; activates the radical-SAM family members Bio2p and Lip5p; interacts with Ccr4p in the two-hybrid system |
|
| YBR144C | 0.89 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR144C is not an essential gene |
||
| YMR063W | 0.89 |
RIM9
|
Protein of unknown function, involved in the proteolytic activation of Rim101p in response to alkaline pH; has similarity to A. nidulans PalI; putative membrane protein |
|
| YBR293W | 0.88 |
VBA2
|
Permease of basic amino acids in the vacuolar membrane |
|
| YGR260W | 0.88 |
TNA1
|
High affinity nicotinic acid plasma membrane permease, responsible for uptake of low levels of nicotinic acid; expression of the gene increases in the absence of extracellular nicotinic acid or para-aminobenzoate (PABA) |
|
| YNL240C | 0.86 |
NAR1
|
Nuclear architecture related protein; component of the cytosolic iron-sulfur (FeS) protein assembly machinery, required for maturation of cytosolic and nuclear FeS proteins; homologous to human Narf |
|
| YPL036W | 0.86 |
PMA2
|
Plasma membrane H+-ATPase, isoform of Pma1p, involved in pumping protons out of the cell; regulator of cytoplasmic pH and plasma membrane potential |
|
| YGL072C | 0.85 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene HSF1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YEL059W | 0.85 |
Dubious open reading frame unlikely to encode a functional protein |
||
| YGR286C | 0.84 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YDL244W | 0.84 |
THI13
|
Protein involved in synthesis of the thiamine precursor hydroxymethylpyrimidine (HMP); member of a subtelomeric gene family including THI5, THI11, THI12, and THI13 |
|
| YGL117W | 0.82 |
Putative protein of unknown function |
||
| YFR047C | 0.81 |
BNA6
|
Quinolinate phosphoribosyl transferase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p |
|
| YBL033C | 0.81 |
RIB1
|
GTP cyclohydrolase II; catalyzes the first step of the riboflavin biosynthesis pathway |
|
| YLL056C | 0.81 |
Putative protein of unknown function, transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in pleiotropic drug resistance (PDR) phenomenon; YLL056C is not an essential gene |
||
| YLR023C | 0.79 |
IZH3
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by zinc deficiency; deletion reduces sensitivity to elevated zinc and shortens lag phase, overexpression reduces Zap1p activity |
|
| YOR227W | 0.77 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YOR336W | 0.77 |
KRE5
|
Protein required for beta-1,6 glucan biosynthesis; mutations result in aberrant morphology and severe growth defects |
|
| YGR142W | 0.76 |
BTN2
|
v-SNARE binding protein that facilitates specific protein retrieval from a late endosome to the Golgi; modulates arginine uptake, possible role in mediating pH homeostasis between the vacuole and plasma membrane H(+)-ATPase |
|
| YOR066W | 0.75 |
MSA1
|
Activator of G1-specific transcription factors, MBF and SBF, that regulates both the timing of G1-specific gene transcription, and cell cycle initiation; potential Cdc28p substrate |
|
| YHR161C | 0.75 |
YAP1801
|
Protein involved in clathrin cage assembly; binds Pan1p and clathrin; homologous to Yap1802p, member of the AP180 protein family |
|
| YKL071W | 0.75 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YGL236C | 0.74 |
MTO1
|
Mitochondrial protein, forms a heterodimer complex with Mss1p that performs the 5-carboxymethylaminomethyl modification of the wobble uridine base in mitochondrial tRNAs; required for respiration in paromomycin-resistant 15S rRNA mutants |
|
| YMR118C | 0.74 |
Protein of unknown function with similarity to succinate dehydrogenase cytochrome b subunit; YMR118C is not an essential gene |
||
| YIR017C | 0.74 |
MET28
|
Transcriptional activator in the Cbf1p-Met4p-Met28p complex, participates in the regulation of sulfur metabolism |
|
| YGL045W | 0.74 |
RIM8
|
Protein of unknown function, involved in the proteolytic activation of Rim101p in response to alkaline pH; has similarity to A. nidulans PalF |
|
| YHR180W | 0.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPR196W | 0.73 |
Putative maltose activator |
||
| YDR542W | 0.73 |
PAU10
|
Hypothetical protein |
|
| YGL180W | 0.72 |
ATG1
|
Protein serine/threonine kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway; structurally required for pre-autophagosome formation; during autophagy forms a complex with Atg13p and Atg17p |
|
| YBR008C | 0.72 |
FLR1
|
Plasma membrane multidrug transporter of the major facilitator superfamily, involved in efflux of fluconazole, diazaborine, benomyl, methotrexate, and other drugs |
|
| YHR139C | 0.71 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YPL033C | 0.71 |
Putative protein of unknown function; may be involved in DNA metabolism; expression is induced by Kar4p |
||
| YLR193C | 0.69 |
UPS1
|
Mitochondrial intermembrane space protein that regulates alternative processing and sorting of Mgm1p and other proteins; required for normal mitochondrial morphology; ortholog of human PRELI |
|
| YJR130C | 0.69 |
STR2
|
Cystathionine gamma-synthase, converts cysteine into cystathionine |
|
| YNL094W | 0.69 |
APP1
|
Protein of unknown function, interacts with Rvs161p and Rvs167p; computational analysis of protein-protein interactions in large-scale studies suggests a possible role in actin filament organization |
|
| YPL264C | 0.68 |
Putative membrane protein of unknown function; physically interacts with Hsp82p; YPL264C is not an essential gene |
||
| YNL103W | 0.68 |
MET4
|
Leucine-zipper transcriptional activator, responsible for the regulation of the sulfur amino acid pathway, requires different combinations of the auxiliary factors Cbf1p, Met28p, Met31p and Met32p |
|
| YMR135C | 0.68 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YPL006W | 0.67 |
NCR1
|
Vacuolar membrane protein that transits through the biosynthetic vacuolar protein sorting pathway, involved in sphingolipid metabolism; glycoprotein and functional orthologue of human Niemann Pick C1 (NPC1) protein |
|
| YHR151C | 0.66 |
MTC6
|
Putative protein of unknown function; YHR151C is synthetically sick with cdc13-1 |
|
| YGL062W | 0.66 |
PYC1
|
Pyruvate carboxylase isoform, cytoplasmic enzyme that converts pyruvate to oxaloacetate; highly similar to isoform Pyc2p but differentially regulated; mutations in the human homolog are associated with lactic acidosis |
|
| YDR525W-A | 0.65 |
SNA2
|
Protein of unknown function, has similarity to Pmp3p, which is involved in cation transport; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YBR292C | 0.65 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YDR034C | 0.65 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YKL167C | 0.64 |
MRP49
|
Mitochondrial ribosomal protein of the large subunit, not essential for mitochondrial translation |
|
| YBL102W | 0.63 |
SFT2
|
Non-essential tetra-spanning membrane protein found mostly in the late Golgi, can suppress some sed5 alleles; may be part of the transport machinery, but precise function is unknown; similar to mammalian syntaxin 5 |
|
| YER066W | 0.63 |
Putative protein of unknown function; YER066W is not an essential gene |
||
| YNL220W | 0.63 |
ADE12
|
Adenylosuccinate synthase, catalyzes the first step in synthesis of adenosine monophosphate from inosine 5'monophosphate during purine nucleotide biosynthesis; exhibits binding to single-stranded autonomously replicating (ARS) core sequence |
|
| YER101C | 0.62 |
AST2
|
Protein that may have a role in targeting of plasma membrane [H+]ATPase (Pma1p) to the plasma membrane, as suggested by analysis of genetic interactions |
|
| YDL218W | 0.62 |
Putative protein of unknown function; YDL218W transcription is regulated by Azf1p and induced by starvation and aerobic conditions |
||
| YLL047W | 0.61 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified gene RNP1 |
||
| YOL117W | 0.61 |
RRI2
|
Subunit of the COP9 signalosome (CSN) complex that cleaves the ubiquitin-like protein Nedd8 from SCF ubiquitin ligases; plays a role in the mating pheromone response |
|
| YDR541C | 0.60 |
Putative dihydrokaempferol 4-reductase |
||
| YEL044W | 0.60 |
IES6
|
Protein that associates with the INO80 chromatin remodeling complex under low-salt conditions |
|
| YKL224C | 0.60 |
PAU16
|
Putative protein of unknown function |
|
| YJL100W | 0.60 |
LSB6
|
Type II phosphatidylinositol 4-kinase that binds Las17p, which is a homolog of human Wiskott-Aldrich Syndrome protein involved in actin patch assembly and actin polymerization |
|
| YMR206W | 0.60 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YMR253C | 0.59 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern; YMR253C is not an essential gene |
||
| YDR476C | 0.59 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; YDR476C is not an essential gene |
||
| YBR035C | 0.59 |
PDX3
|
Pyridoxine (pyridoxamine) phosphate oxidase, has homologs in E. coli and Myxococcus xanthus; transcription is under the general control of nitrogen metabolism |
|
| YBR037C | 0.59 |
SCO1
|
Copper-binding protein of the mitochondrial inner membrane, required for cytochrome c oxidase activity and respiration; may function to deliver copper to cytochrome c oxidase; has similarity to thioredoxins |
|
| YPL230W | 0.59 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YGL255W | 0.58 |
ZRT1
|
High-affinity zinc transporter of the plasma membrane, responsible for the majority of zinc uptake; transcription is induced under low-zinc conditions by the Zap1p transcription factor |
|
| YNL095C | 0.57 |
Putative protein of unknown function predicted to contain a transmembrane domain; YNL095C is not an essential gene |
||
| YGR087C | 0.56 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YOR255W | 0.56 |
OSW1
|
Protein involved in sporulation; required for the construction of the outer spore wall layers; required for proper localization of Spo14p |
|
| YPR027C | 0.56 |
Putative protein of unknown function |
||
| YDL194W | 0.56 |
SNF3
|
Plasma membrane glucose sensor that regulates glucose transport; has 12 predicted transmembrane segments; long cytoplasmic C-terminal tail is required for low glucose induction of hexose transporter genes HXT2 and HXT4 |
|
| YBR105C | 0.55 |
VID24
|
Peripheral membrane protein located at Vid (vacuole import and degradation) vesicles; regulates fructose-1,6-bisphosphatase (FBPase) targeting to the vacuole; involved in proteasome-dependent catabolite degradation of FBPase |
|
| YOR346W | 0.55 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YGR273C | 0.54 |
Putative protein of unknown function; deletion mutant has no readily detectable phenotype; expression downregulated by treatment with 8-methoxypsoralen plus UVA irradiation |
||
| YFL025C | 0.54 |
BST1
|
GPI inositol deacylase of the ER that negatively regulates COPII vesicle formation, prevents production of vesicles with defective subunits, required for proper discrimination between resident ER proteins and Golgi-bound cargo molecules |
|
| YJL089W | 0.54 |
SIP4
|
C6 zinc cluster transcriptional activator that binds to the carbon source-responsive element (CSRE) of gluconeogenic genes; involved in the positive regulation of gluconeogenesis; regulated by Snf1p protein kinase; localized to the nucleus |
|
| YIL060W | 0.54 |
Putative protein of unknown function; mutant accumulates less glycogen than does wild type; YIL060W is not an essential gene |
||
| YPL183W-A | 0.54 |
RTC6
|
Homolog of the prokaryotic ribosomal protein L36, likely to be a mitochondrial ribosomal protein coded in the nuclear genome; null mutation suppresses cdc13-1 temperature sensitivity |
|
| YLR228C | 0.54 |
ECM22
|
Sterol regulatory element binding protein, regulates transcription of the sterol biosynthetic genes ERG2 and ERG3; member of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors; homologous to Upc2p |
|
| YCL048W | 0.53 |
SPS22
|
Protein of unknown function, redundant with Sps2p for the organization of the beta-glucan layer of the spore wall |
|
| YBL049W | 0.53 |
MOH1
|
Protein of unknown function, has homology to kinase Snf7p; not required for growth on nonfermentable carbon sources; essential for viability in stationary phase |
|
| YIR039C | 0.53 |
YPS6
|
Putative GPI-anchored aspartic protease |
|
| YPL229W | 0.53 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YPL229W is not an essential gene |
||
| YJL170C | 0.53 |
ASG7
|
Protein that regulates signaling from a G protein beta subunit Ste4p and its relocalization within the cell; specific to a-cells and induced by alpha-factor |
|
| YIR018W | 0.52 |
YAP5
|
Basic leucine zipper (bZIP) transcription factor |
|
| YGL170C | 0.52 |
SPO74
|
Component of the meiotic outer plaque of the spindle pole body, involved in modifying the meiotic outer plaque that is required prior to prospore membrane formation |
|
| YOR192C | 0.52 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YOL026C | 0.50 |
MIM1
|
Mitochondrial outer membrane protein, required for assembly of the translocase of the outer membrane (TOM) complex and thereby for mitochondrial protein import; N terminus is exposed to the cytosol: transmembrane segment is highly conserved |
|
| YPL017C | 0.50 |
IRC15
|
Putative S-adenosylmethionine-dependent methyltransferase of the seven beta-strand family, required for accurate meiotic chromosome segregation; null mutant displays increased levels of spontaneous Rad52 foci |
|
| YGL141W | 0.50 |
HUL5
|
Ubiquitin-conjugating enzyme (E4), elongates polyubiquitin chains on substrate proteins; works in opposition to Ubp6p polyubiquitin-shortening activity; required for retrograde transport of misfolded proteins into cytoplasm during ERAD |
|
| YKR098C | 0.50 |
UBP11
|
Ubiquitin-specific protease that cleaves ubiquitin from ubiquitinated proteins |
|
| YMR170C | 0.49 |
ALD2
|
Cytoplasmic aldehyde dehydrogenase, involved in ethanol oxidation and beta-alanine biosynthesis; uses NAD+ as the preferred coenzyme; expression is stress induced and glucose repressed; very similar to Ald3p |
|
| YLR058C | 0.49 |
SHM2
|
Cytosolic serine hydroxymethyltransferase, converts serine to glycine plus 5,10 methylenetetrahydrofolate; major isoform involved in generating precursors for purine, pyrimidine, amino acid, and lipid biosynthesis |
|
| YJL210W | 0.48 |
PEX2
|
RING-finger peroxin, peroxisomal membrane protein with a C-terminal zinc-binding RING domain, forms translocation subcomplex with Pex10p and Pex12p which functions in peroxisomal matrix protein import |
Network of associatons between targets according to the STRING database.
First level regulatory network of GCN4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 1.2 | 9.9 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.9 | 3.5 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.8 | 3.2 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.7 | 7.4 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.7 | 5.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.7 | 1.3 | GO:0045117 | azole transport(GO:0045117) |
| 0.7 | 2.0 | GO:0015755 | fructose transport(GO:0015755) |
| 0.7 | 4.6 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.6 | 1.9 | GO:0009437 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.6 | 2.9 | GO:0043433 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.5 | 2.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.5 | 2.7 | GO:0044746 | amino acid transmembrane export from vacuole(GO:0032974) vacuolar transmembrane transport(GO:0034486) vacuolar amino acid transmembrane transport(GO:0034487) amino acid transmembrane export(GO:0044746) |
| 0.5 | 1.5 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.5 | 2.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.5 | 7.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.5 | 1.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.5 | 1.4 | GO:0034395 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.5 | 1.4 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.4 | 1.8 | GO:0019748 | secondary metabolic process(GO:0019748) |
| 0.4 | 3.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 1.3 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.4 | 3.2 | GO:0005987 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.4 | 1.5 | GO:0043200 | response to amino acid(GO:0043200) |
| 0.4 | 3.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 2.4 | GO:0034762 | regulation of transmembrane transport(GO:0034762) |
| 0.3 | 7.5 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.3 | 1.8 | GO:0000501 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.3 | 0.6 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.3 | 0.5 | GO:0061414 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061413) positive regulation of transcription from RNA polymerase II promoter by a nonfermentable carbon source(GO:0061414) |
| 0.2 | 1.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.2 | 3.8 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.2 | 1.2 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.2 | 0.9 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.2 | 1.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.2 | 1.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.2 | 2.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.2 | 0.6 | GO:0070583 | spore membrane bending pathway(GO:0070583) |
| 0.2 | 0.6 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.2 | 1.0 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 0.6 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.2 | 2.6 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.2 | 0.6 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.2 | 1.6 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 1.2 | GO:0015891 | siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:0051238 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.5 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.1 | 2.5 | GO:0008645 | hexose transport(GO:0008645) monosaccharide transport(GO:0015749) |
| 0.1 | 0.5 | GO:0000200 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.1 | 0.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.4 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.1 | 1.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.2 | GO:2001038 | cellular response to drug(GO:0035690) regulation of response to drug(GO:2001023) positive regulation of response to drug(GO:2001025) regulation of cellular response to drug(GO:2001038) positive regulation of cellular response to drug(GO:2001040) |
| 0.1 | 0.6 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 3.3 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.5 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.4 | GO:0000435 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.1 | 0.4 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.1 | 0.5 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 1.0 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.1 | 0.5 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 0.2 | GO:0001113 | transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) |
| 0.1 | 3.0 | GO:0071940 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.1 | 0.3 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.1 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.1 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.1 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061395) cellular response to arsenic-containing substance(GO:0071243) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.2 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.1 | 0.3 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 | 1.9 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.2 | GO:0001109 | promoter clearance during DNA-templated transcription(GO:0001109) promoter clearance from RNA polymerase II promoter(GO:0001111) |
| 0.1 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.9 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.3 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.4 | GO:0000105 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.0 | 0.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:0000755 | cytogamy(GO:0000755) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0043954 | cellular component maintenance(GO:0043954) |
| 0.0 | 0.0 | GO:0010446 | response to alkaline pH(GO:0010446) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0019935 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 0.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.5 | GO:0015849 | organic acid transport(GO:0015849) |
| 0.0 | 0.1 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.0 | 0.1 | GO:0030491 | heteroduplex formation(GO:0030491) |
| 0.0 | 0.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0000436 | carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0000348 | mRNA branch site recognition(GO:0000348) |
| 0.0 | 0.2 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 1.0 | GO:0030437 | ascospore formation(GO:0030437) cell development(GO:0048468) |
| 0.0 | 0.2 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.4 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.1 | GO:0006784 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 2.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0031503 | protein complex localization(GO:0031503) |
| 0.0 | 0.1 | GO:0019405 | alditol catabolic process(GO:0019405) glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.0 | 0.2 | GO:0000147 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.1 | GO:0045833 | negative regulation of lipid metabolic process(GO:0045833) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.8 | 2.5 | GO:0000274 | mitochondrial proton-transporting ATP synthase, stator stalk(GO:0000274) proton-transporting ATP synthase, stator stalk(GO:0045265) |
| 0.6 | 2.5 | GO:0000817 | COMA complex(GO:0000817) |
| 0.3 | 1.7 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 2.7 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.3 | 0.8 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.2 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.0 | GO:0044233 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.0 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.1 | 0.3 | GO:0032126 | eisosome(GO:0032126) |
| 0.1 | 1.8 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.3 | GO:0031160 | spore wall(GO:0031160) |
| 0.1 | 3.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.4 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.2 | GO:0042764 | prospore membrane(GO:0005628) intracellular immature spore(GO:0042763) ascospore-type prospore(GO:0042764) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 22.5 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.1 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.3 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 2.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0044284 | mitochondrial crista junction(GO:0044284) |
| 0.0 | 0.7 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0070274 | RES complex(GO:0070274) |
| 0.0 | 0.1 | GO:0098562 | side of membrane(GO:0098552) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.2 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.2 | GO:0071013 | U2-type catalytic step 2 spliceosome(GO:0071007) catalytic step 2 spliceosome(GO:0071013) post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.0 | 0.1 | GO:0000799 | condensin complex(GO:0000796) nuclear condensin complex(GO:0000799) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0004575 | beta-fructofuranosidase activity(GO:0004564) sucrose alpha-glucosidase activity(GO:0004575) |
| 0.9 | 2.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.7 | 3.0 | GO:0015603 | siderophore transmembrane transporter activity(GO:0015343) iron chelate transmembrane transporter activity(GO:0015603) siderophore transporter activity(GO:0042927) |
| 0.7 | 8.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.7 | 2.2 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.7 | 3.5 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.6 | 1.9 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) carnitine O-acyltransferase activity(GO:0016406) |
| 0.6 | 4.8 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.5 | 2.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.5 | 2.1 | GO:0015146 | pentose transmembrane transporter activity(GO:0015146) |
| 0.5 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 2.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.5 | 2.9 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.5 | 1.4 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.5 | 3.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.4 | 4.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.4 | 2.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.4 | 2.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.4 | 3.2 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.4 | 1.6 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 1.4 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.4 | 0.7 | GO:0045118 | azole transporter activity(GO:0045118) |
| 0.3 | 0.3 | GO:0015489 | putrescine transmembrane transporter activity(GO:0015489) |
| 0.3 | 1.0 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 9.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.3 | 1.4 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.3 | 2.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 3.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.7 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.2 | 1.0 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 0.6 | GO:0005536 | glucose binding(GO:0005536) |
| 0.2 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.7 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.2 | 0.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.6 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.6 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 5.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.3 | GO:0015662 | ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism(GO:0015662) |
| 0.1 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.8 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.2 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.3 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.1 | 0.1 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.1 | 0.9 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.1 | 0.9 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) NADH kinase activity(GO:0042736) |
| 0.1 | 1.5 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 0.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.4 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 0.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 2.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.1 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.8 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 0.5 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.1 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.2 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.1 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.6 | GO:0038023 | receptor activity(GO:0004872) transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) molecular transducer activity(GO:0060089) transmembrane receptor activity(GO:0099600) |
| 0.1 | 0.3 | GO:0042123 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.1 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.2 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.1 | 0.2 | GO:0004458 | lactate dehydrogenase activity(GO:0004457) D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.6 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.4 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.4 | GO:0015578 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.0 | 0.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.4 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.3 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 56.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.3 | 3.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 56.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |