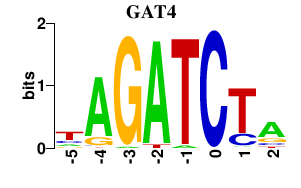
Results for GAT4
Z-value: 0.73
Transcription factors associated with GAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GAT4
|
S000001452 | Protein containing GATA family zinc finger motifs |
Activity-expression correlation:
Activity profile of GAT4 motif
Sorted Z-values of GAT4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YIL057C | 7.44 |
Putative protein of unknown function; expression induced under carbon limitation and repressed under high glucose |
||
| YPL223C | 5.24 |
GRE1
|
Hydrophilin of unknown function; stress induced (osmotic, ionic, oxidative, heat shock and heavy metals); regulated by the HOG pathway |
|
| YKR097W | 4.48 |
PCK1
|
Phosphoenolpyruvate carboxykinase, key enzyme in gluconeogenesis, catalyzes early reaction in carbohydrate biosynthesis, glucose represses transcription and accelerates mRNA degradation, regulated by Mcm1p and Cat8p, located in the cytosol |
|
| YAR053W | 4.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR139C | 4.21 |
SPS100
|
Protein required for spore wall maturation; expressed during sporulation; may be a component of the spore wall |
|
| YKL217W | 4.17 |
JEN1
|
Lactate transporter, required for uptake of lactate and pyruvate; phosphorylated; expression is derepressed by transcriptional activator Cat8p during respiratory growth, and repressed in the presence of glucose, fructose, and mannose |
|
| YOR343C | 3.19 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YNL093W | 3.15 |
YPT53
|
GTPase, similar to Ypt51p and Ypt52p and to mammalian rab5; required for vacuolar protein sorting and endocytosis |
|
| YJR150C | 3.09 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YJR146W | 3.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene HMS2 |
||
| YHR212W-A | 2.90 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YHR212C | 2.87 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL177W | 2.78 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR236C | 2.75 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YBR072W | 2.61 |
HSP26
|
Small heat shock protein (sHSP) with chaperone activity; forms hollow, sphere-shaped oligomers that suppress unfolded proteins aggregation; oligomer activation requires a heat-induced conformational change; not expressed in unstressed cells |
|
| YPL054W | 2.60 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YAR060C | 2.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR096C | 2.45 |
HXT5
|
Hexose transporter with moderate affinity for glucose, induced in the presence of non-fermentable carbon sources, induced by a decrease in growth rate, contains an extended N-terminal domain relative to other HXTs |
|
| YJL045W | 2.45 |
Minor succinate dehydrogenase isozyme; homologous to Sdh1p, the major isozyme reponsible for the oxidation of succinate and transfer of electrons to ubiquinone; induced during the diauxic shift in a Cat8p-dependent manner |
||
| YML128C | 2.35 |
MSC1
|
Protein of unknown function; mutant is defective in directing meiotic recombination events to homologous chromatids; the authentic, non-tagged protein is detected in highly purified mitochondria and is phosphorylated |
|
| YGR067C | 2.30 |
Putative protein of unknown function; contains a zinc finger motif similar to that of Adr1p |
||
| YEL039C | 2.27 |
CYC7
|
Cytochrome c isoform 2, expressed under hypoxic conditions; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YLR311C | 2.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YEL070W | 2.25 |
DSF1
|
Deletion suppressor of mpt5 mutation |
|
| YOR178C | 2.22 |
GAC1
|
Regulatory subunit for Glc7p type-1 protein phosphatase (PP1), tethers Glc7p to Gsy2p glycogen synthase, binds Hsf1p heat shock transcription factor, required for induction of some HSF-regulated genes under heat shock |
|
| YPL156C | 2.15 |
PRM4
|
Pheromone-regulated protein, predicted to have 1 transmembrane segment; transcriptionally regulated by Ste12p during mating and by Cat8p during the diauxic shift |
|
| YPR030W | 2.08 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YPL222W | 1.99 |
FMP40
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YPR013C | 1.99 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YPR018W | 1.98 |
RLF2
|
Largest subunit (p90) of the Chromatin Assembly Complex (CAF-1) with Cac2p and Msi1p that assembles newly synthesized histones onto recently replicated DNA; involved in the maintenance of transcriptionally silent chromatin |
|
| YAR019W-A | 1.97 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR393W | 1.97 |
ERR1
|
Protein of unknown function, has similarity to enolases |
|
| YAL062W | 1.95 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YAL054C | 1.91 |
ACS1
|
Acetyl-coA synthetase isoform which, along with Acs2p, is the nuclear source of acetyl-coA for histone acetlyation; expressed during growth on nonfermentable carbon sources and under aerobic conditions |
|
| YMR191W | 1.90 |
SPG5
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YAR050W | 1.89 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YKR011C | 1.88 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YGR039W | 1.82 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YGR022C | 1.78 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF MTL1/YGR023W |
||
| YGR023W | 1.76 |
MTL1
|
Protein with both structural and functional similarity to Mid2p, which is a plasma membrane sensor required for cell integrity signaling during pheromone-induced morphogenesis; suppresses rgd1 null mutations |
|
| YMR316C-B | 1.75 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YFR053C | 1.74 |
HXK1
|
Hexokinase isoenzyme 1, a cytosolic protein that catalyzes phosphorylation of glucose during glucose metabolism; expression is highest during growth on non-glucose carbon sources; glucose-induced repression involves the hexokinase Hxk2p |
|
| YEL030C-A | 1.70 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YNR034W-A | 1.69 |
Putative protein of unknown function; expression is regulated by Msn2p/Msn4p |
||
| YPR169W-A | 1.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YBR108W | 1.64 |
AIM3
|
Protein interacting with Rsv167p; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YAR019C | 1.57 |
CDC15
|
Protein kinase of the Mitotic Exit Network that is localized to the spindle pole bodies at late anaphase; promotes mitotic exit by directly switching on the kinase activity of Dbf2p |
|
| YER093C-A | 1.56 |
AIM11
|
Putative protein of unknown function; YER093C-A contains an intron; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YPR061C | 1.55 |
JID1
|
Probable Hsp40p co-chaperone, has a DnaJ-like domain and appears to be involved in ER-associated degradation of misfolded proteins containing a tightly folded cytoplasmic domain; inhibits replication of Brome mosaic virus in S. cerevisiae |
|
| YMR317W | 1.55 |
Putative protein of unknown function with some similarity to sialidase from Trypanosoma; YMR317W is not an essential gene |
||
| YEL030W | 1.52 |
ECM10
|
Heat shock protein of the Hsp70 family, localized in mitochondrial nucleoids, plays a role in protein translocation, interacts with Mge1p in an ATP-dependent manner; overexpression induces extensive mitochondrial DNA aggregations |
|
| YGL138C | 1.52 |
Putative protein of unknown function; has no significant sequence similarity to any known protein |
||
| YNL036W | 1.51 |
NCE103
|
Carbonic anhydrase; poorly transcribed under aerobic conditions and at an undetectable level under anaerobic conditions; involved in non-classical protein export pathway |
|
| YOR391C | 1.51 |
HSP33
|
Possible chaperone and cysteine protease with similarity to E. coli Hsp31 and S. cerevisiae Hsp31p, Hsp32p, and Sno4p; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease |
|
| YDR018C | 1.50 |
Probable membrane protein with three predicted transmembrane domains; homologous to Ybr042cp, similar to C. elegans F55A11.5 and maize 1-acyl-glycerol-3-phosphate acyltransferase; null exhibits no apparent phenotype |
||
| YDR540C | 1.49 |
IRC4
|
Putative protein of unknown function; null mutant displays increased levels of spontaneous Rad52p foci; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
|
| YGR286C | 1.49 |
BIO2
|
Biotin synthase, catalyzes the conversion of dethiobiotin to biotin, which is the last step of the biotin biosynthesis pathway; complements E. coli bioB mutant |
|
| YER147C | 1.47 |
SCC4
|
Subunit of cohesin loading factor (Scc2p-Scc4p), a complex required for the loading of cohesin complexes onto chromosomes; involved in establishing sister chromatid cohesion during double-strand break repair via phosphorylated histone H2AX |
|
| YLL019C | 1.43 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YLR332W | 1.43 |
MID2
|
O-glycosylated plasma membrane protein that acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with cell integrity pathway protein Zeo1p |
|
| YGR070W | 1.42 |
ROM1
|
GDP/GTP exchange protein (GEP) for Rho1p; mutations are synthetically lethal with mutations in rom2, which also encodes a GEP |
|
| YML087C | 1.41 |
AIM33
|
Putative protein of unknown function; highly conserved across species and orthologous to human CYB5R4; null mutant shows increased frequency of mitochondrial genome loss (petite formation) and severe growth defect in minimal glycerol media |
|
| YJR151C | 1.41 |
DAN4
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YDR077W | 1.40 |
SED1
|
Major stress-induced structural GPI-cell wall glycoprotein in stationary-phase cells, associates with translating ribosomes, possible role in mitochondrial genome maintenance; ORF contains two distinct variable minisatellites |
|
| YAR047C | 1.39 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YHR126C | 1.36 |
Putative protein of unknown function; transcription dependent upon Azf1p |
||
| YOR348C | 1.36 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YJR152W | 1.36 |
DAL5
|
Allantoin permease; ureidosuccinate permease; expression is constitutive but sensitive to nitrogen catabolite repression |
|
| YIL045W | 1.35 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YKR102W | 1.33 |
FLO10
|
Lectin-like protein with similarity to Flo1p, thought to be involved in flocculation |
|
| YGR213C | 1.32 |
RTA1
|
Protein involved in 7-aminocholesterol resistance; has seven potential membrane-spanning regions; expression is induced under both low-heme and low-oxygen conditions |
|
| YNL052W | 1.32 |
COX5A
|
Subunit Va of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; predominantly expressed during aerobic growth while its isoform Vb (Cox5Bp) is expressed during anaerobic growth |
|
| YLR331C | 1.30 |
JIP3
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene MID2 |
|
| YMR104C | 1.25 |
YPK2
|
Protein kinase with similarityto serine/threonine protein kinase Ypk1p; functionally redundant with YPK1 at the genetic level; participates in a signaling pathway required for optimal cell wall integrity; homolog of mammalian kinase SGK |
|
| YDR034C | 1.25 |
LYS14
|
Transcriptional activator involved in regulation of genes of the lysine biosynthesis pathway; requires 2-aminoadipate semialdehyde as co-inducer |
|
| YBR076C-A | 1.24 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YLR312C | 1.21 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YLR037C | 1.21 |
DAN2
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YLR228C | 1.20 |
ECM22
|
Sterol regulatory element binding protein, regulates transcription of the sterol biosynthetic genes ERG2 and ERG3; member of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors; homologous to Upc2p |
|
| YIL024C | 1.19 |
Putative protein of unknown function; non-essential gene; expression directly regulated by the metabolic and meiotic transcriptional regulator Ume6p |
||
| YKR101W | 1.18 |
SIR1
|
Protein involved in repression of transcription at the silent mating-type loci HML and HMR; recruitment to silent chromatin requires interactions with Orc1p and with Sir4p, through a common Sir1p domain; binds to centromeric chromatin |
|
| YNL180C | 1.18 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YLR247C | 1.18 |
IRC20
|
Putative helicase; localizes to the mitochondrion and the nucleus; YLR247C is not an essential gene; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YDR277C | 1.17 |
MTH1
|
Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation |
|
| YJR048W | 1.17 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YJL116C | 1.16 |
NCA3
|
Protein that functions with Nca2p to regulate mitochondrial expression of subunits 6 (Atp6p) and 8 (Atp8p ) of the Fo-F1 ATP synthase; member of the SUN family |
|
| YOL118C | 1.15 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YKR100C | 1.15 |
SKG1
|
Transmembrane protein with a role in cell wall polymer composition; localizes on the inner surface of the plasma membrane at the bud and in the daughter cell |
|
| YNL144C | 1.14 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YNL144C is not an essential gene |
||
| YCR010C | 1.14 |
ADY2
|
Acetate transporter required for normal sporulation; phosphorylated in mitochondria |
|
| YNL125C | 1.13 |
ESBP6
|
Protein with similarity to monocarboxylate permeases, appears not to be involved in transport of monocarboxylates such as lactate, pyruvate or acetate across the plasma membrane |
|
| YDL085W | 1.13 |
NDE2
|
Mitochondrial external NADH dehydrogenase, catalyzes the oxidation of cytosolic NADH; Nde1p and Nde2p are involved in providing the cytosolic NADH to the mitochondrial respiratory chain |
|
| YGL089C | 1.13 |
MF(ALPHA)2
|
Mating pheromone alpha-factor, made by alpha cells; interacts with mating type a cells to induce cell cycle arrest and other responses leading to mating; also encoded by MF(ALPHA)1, which is more highly expressed than MF(ALPHA)2 |
|
| YJR078W | 1.10 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YDR536W | 1.10 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YDR322C-A | 1.08 |
TIM11
|
Subunit e of mitochondrial F1F0-ATPase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis; essential for the dimeric and oligomeric state of ATP synthase |
|
| YJR094C | 1.08 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YNL179C | 1.08 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YLR307C-A | 1.05 |
Putative protein of unknown function |
||
| YAL067C | 1.04 |
SEO1
|
Putative permease, member of the allantoate transporter subfamily of the major facilitator superfamily; mutation confers resistance to ethionine sulfoxide |
|
| YOR186W | 1.04 |
Putative protein of unknown function; proper regulation of expression during heat stress is sphingolipid-dependent |
||
| YOL055C | 1.04 |
THI20
|
Multifunctional protein with both hydroxymethylpyrimidine kinase and thiaminase activities; involved in thiamine biosynthesis and also in thiamine degradation; member of a gene family with THI21 and THI22; functionally redundant with Thi21p |
|
| YGL163C | 1.01 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YPL018W | 1.00 |
CTF19
|
Outer kinetochore protein, required for accurate mitotic chromosome segregation; component of the kinetochore sub-complex COMA (Ctf19p, Okp1p, Mcm21p, Ame1p) that functions as a platform for kinetochore assembly |
|
| YOR208W | 0.99 |
PTP2
|
Phosphotyrosine-specific protein phosphatase involved in the inactivation of mitogen-activated protein kinase (MAPK) during osmolarity sensing; dephosporylates Hog1p MAPK and regulates its localization; localized to the nucleus |
|
| YIL036W | 0.99 |
CST6
|
Basic leucine zipper (bZIP) transcription factor of the ATF/CREB family, activates transcription of genes involved in utilization of non-optimal carbon sources; involved in telomere maintenance |
|
| YNL091W | 0.98 |
NST1
|
Protein of unknown function, mediates sensitivity to salt stress; interacts physically with the splicing factor Msl1p and also displays genetic interaction with MSL1 |
|
| YBR077C | 0.97 |
SLM4
|
Component of the EGO complex, which is involved in the regulation of microautophagy, and of the GSE complex, which is required for proper sorting of amino acid permease Gap1p; gene exhibits synthetic genetic interaction with MSS4 |
|
| YPR191W | 0.97 |
QCR2
|
Subunit 2 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; phosphorylated; transcription is regulated by Hap1p, Hap2p/Hap3p, and heme |
|
| YIL037C | 0.96 |
PRM2
|
Pheromone-regulated protein, predicted to have 4 transmembrane segments and a coiled coil domain; regulated by Ste12p |
|
| YNL074C | 0.95 |
MLF3
|
Serine-rich protein of unknown function; overproduction suppresses the growth inhibition caused by exposure to the immunosuppressant leflunomide |
|
| YMR201C | 0.92 |
RAD14
|
Protein that recognizes and binds damaged DNA during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 1 (NEF1); contains zinc finger motif; homolog of human XPA protein |
|
| YAL034C | 0.92 |
FUN19
|
Non-essential protein of unknown function |
|
| YMR206W | 0.92 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YFR017C | 0.92 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and is induced in response to the DNA-damaging agent MMS; YFR017C is not an essential gene |
||
| YCR021C | 0.91 |
HSP30
|
Hydrophobic plasma membrane localized, stress-responsive protein that negatively regulates the H(+)-ATPase Pma1p; induced by heat shock, ethanol treatment, weak organic acid, glucose limitation, and entry into stationary phase |
|
| YHR139C-A | 0.90 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR223C | 0.89 |
IFH1
|
Essential protein with a highly acidic N-terminal domain; IFH1 exhibits genetic interactions with FHL1, overexpression interferes with silencing at telomeres and HM loci; potential Cdc28p substrate |
|
| YIL122W | 0.89 |
POG1
|
Putative transcriptional activator that promotes recovery from pheromone induced arrest; inhibits both alpha-factor induced G1 arrest and repression of CLN1 and CLN2 via SCB/MCB promoter elements; potential Cdc28p substrate; SBF regulated |
|
| YJR115W | 0.88 |
Putative protein of unknown function |
||
| YPL058C | 0.86 |
PDR12
|
Plasma membrane ATP-binding cassette (ABC) transporter, weak-acid-inducible multidrug transporter required for weak organic acid resistance; induced by sorbate and benzoate and regulated by War1p; mutants exhibit sorbate hypersensitivity |
|
| YOR345C | 0.86 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YEL012W | 0.86 |
UBC8
|
Ubiquitin-conjugating enzyme that negatively regulates gluconeogenesis by mediating the glucose-induced ubiquitination of fructose-1,6-bisphosphatase (FBPase); cytoplasmic enzyme that catalyzes the ubiquitination of histones in vitro |
|
| YJL135W | 0.85 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified genes YJL134W/LCB3 |
||
| YOR346W | 0.85 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YFL052W | 0.84 |
Putative zinc cluster protein that contains a DNA binding domain; null mutant sensitive to calcofluor white, low osmolarity and heat, suggesting a role for YFL052Wp in cell wall integrity |
||
| YNL194C | 0.84 |
Integral membrane protein localized to eisosomes; sporulation and plasma membrane sphingolipid content are altered in mutants; has homologs SUR7 and FMP45; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YDR446W | 0.83 |
ECM11
|
Non-essential protein apparently involved in meiosis, GFP fusion protein is present in discrete clusters in the nucleus throughout mitosis; may be involved in maintaining chromatin structure |
|
| YNL142W | 0.83 |
MEP2
|
Ammonium permease involved in regulation of pseudohyphal growth; belongs to a ubiquitous family of cytoplasmic membrane proteins that transport only ammonium (NH4+); expression is under the nitrogen catabolite repression regulation |
|
| YBR132C | 0.83 |
AGP2
|
High affinity polyamine permease, preferentially uses spermidine over putrescine; expression is down-regulated by osmotic stress; plasma membrane carnitine transporter, also functions as a low-affinity amino acid permease |
|
| YDR119W-A | 0.82 |
Putative protein of unknown function |
||
| YBL039W-B | 0.81 |
Putative protein of unknown function |
||
| YFR023W | 0.81 |
PES4
|
Poly(A) binding protein, suppressor of DNA polymerase epsilon mutation, similar to Mip6p |
|
| YNL143C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YCR007C | 0.80 |
Putative integral membrane protein, member of DUP240 gene family; YCR007C is not an essential gene |
||
| YDR350C | 0.79 |
ATP22
|
Mitochondrial inner membrane protein required for assembly of the F0 sector of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis |
|
| YER103W | 0.79 |
SSA4
|
Heat shock protein that is highly induced upon stress; plays a role in SRP-dependent cotranslational protein-membrane targeting and translocation; member of the HSP70 family; cytoplasmic protein that concentrates in nuclei upon starvation |
|
| YDL224C | 0.78 |
WHI4
|
Putative RNA binding protein and partially redundant Whi3p homolog that regulates the cell size requirement for passage through Start and commitment to cell division |
|
| YIL013C | 0.77 |
PDR11
|
ATP-binding cassette (ABC) transporter, multidrug transporter involved in multiple drug resistance; mediates sterol uptake when sterol biosynthesis is compromisedregulated by Pdr1p; required for anaerobic growth |
|
| YDR055W | 0.77 |
PST1
|
Cell wall protein that contains a putative GPI-attachment site; secreted by regenerating protoplasts; up-regulated by activation of the cell integrity pathway, as mediated by Rlm1p; upregulated by cell wall damage via disruption of FKS1 |
|
| YLR390W-A | 0.76 |
CCW14
|
Covalently linked cell wall glycoprotein, present in the inner layer of the cell wall |
|
| YIL147C | 0.76 |
SLN1
|
Histidine kinase osmosensor that regulates a MAP kinase cascade; transmembrane protein with an intracellular kinase domain that signals to Ypd1p and Ssk1p, thereby forming a phosphorelay system similar to bacterial two-component regulators |
|
| YNL176C | 0.75 |
Cell cycle-regulated gene of unknown function, promoter bound by Fkh2p |
||
| YJL057C | 0.74 |
IKS1
|
Putative serine/threonine kinase; expression is induced during mild heat stress; deletion mutants are hypersensitive to copper sulphate and resistant to sorbate; interacts with an N-terminal fragment of Sst2p |
|
| YIL146C | 0.74 |
ECM37
|
Non-essential protein of unknown function |
|
| YIL047C | 0.74 |
SYG1
|
Plasma membrane protein of unknown function; truncation and overexpression suppresses lethality of G-alpha protein deficiency |
|
| YMR135C | 0.74 |
GID8
|
Protein of unknown function, involved in proteasome-dependent catabolite inactivation of fructose-1,6-bisphosphatase; contains LisH and CTLH domains, like Vid30p; dosage-dependent regulator of START |
|
| YNR047W | 0.73 |
Putative protein kinase that, when overexpressed, interferes with pheromone-induced growth arrest; localizes to the cytoplasm; potential Cdc28p substrate |
||
| YMR040W | 0.72 |
YET2
|
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YPL116W | 0.72 |
HOS3
|
Trichostatin A-insensitive homodimeric histone deacetylase (HDAC) with specificity in vitro for histones H3, H4, H2A, and H2B; similar to Hda1p, Rpd3p, Hos1p, and Hos2p; deletion results in increased histone acetylation at rDNA repeats |
|
| YGL114W | 0.71 |
Putative protein of unknown function; predicted member of the oligopeptide transporter (OPT) family of membrane transporters |
||
| YMR020W | 0.71 |
FMS1
|
Polyamine oxidase, converts spermine to spermidine, which is required for the essential hypusination modification of translation factor eIF-5A; also involved in pantothenic acid biosynthesis |
|
| YGL096W | 0.70 |
TOS8
|
Homeodomain-containing transcription factor; SBF regulated target gene that in turn regulates expression of genes involved in G1/S phase events such as bud site selection, bud emergence and cell cycle progression; similarity to Cup9p |
|
| YDR351W | 0.70 |
SBE2
|
Protein involved in the transport of cell wall components from the Golgi to the cell surface; required for bud growth |
|
| YDR515W | 0.69 |
SLF1
|
RNA binding protein that associates with polysomes; proposed to be involved in regulating mRNA translation; involved in the copper-dependent mineralization of copper sulfide complexes on cell surface in cells cultured in copper salts |
|
| YNL037C | 0.69 |
IDH1
|
Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle |
|
| YER014C-A | 0.68 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YGR248W | 0.68 |
SOL4
|
6-phosphogluconolactonase with similarity to Sol3p |
|
| YOR392W | 0.68 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; gene expression induced by heat |
||
| YMR030W | 0.68 |
RSF1
|
Protein required for respiratory growth; localized to both the nucleus and mitochondrion; mutant displays decreased transcription of specific nuclear and mitochondrial genes whose products are involved in respiratory growth |
|
| YNL223W | 0.67 |
ATG4
|
Conserved cysteine protease required for autophagy; cleaves Atg8p to a form required for autophagosome and Cvt vesicle generation; mediates attachment of autophagosomes to microtubules through interactions with Tub1p and Tub2p |
|
| YER167W | 0.67 |
BCK2
|
Protein rich in serine and threonine residues involved in protein kinase C signaling pathway, which controls cell integrity; overproduction suppresses pkc1 mutations |
|
| YGR069W | 0.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL089C | 0.67 |
RLM1
|
MADS-box transcription factor, component of the protein kinase C-mediated MAP kinase pathway involved in the maintenance of cell integrity; phosphorylated and activated by the MAP-kinase Slt2p |
|
| YKL071W | 0.67 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm |
||
| YLR189C | 0.66 |
ATG26
|
UDP-glucose:sterol glucosyltransferase, conserved enzyme involved in synthesis of sterol glucoside membrane lipids; in contrast to ATG26 from P. pastoris, S. cerevisiae ATG26 is not involved in autophagy |
|
| YJR160C | 0.66 |
MPH3
|
Alpha-glucoside permease, transports maltose, maltotriose, alpha-methylglucoside, and turanose; identical to Mph2p; encoded in a subtelomeric position in a region likely to have undergone duplication |
|
| YMR190C | 0.65 |
SGS1
|
Nucleolar DNA helicase of the RecQ family involved in maintenance of genome integrity, regulates chromosome synapsis and meiotic crossing over; has similarity to human BLM and WRN helicases implicated in Bloom and Werner syndromes |
|
| YLR387C | 0.65 |
REH1
|
Protein of unknown function, similar to Rei1p but not involved in bud growth; contains dispersed C2H2 zinc finger domains |
|
| YMR273C | 0.65 |
ZDS1
|
Protein that interacts with silencing proteins at the telomere, involved in transcriptional silencing; has a role in localization of Bcy1p, a regulatory subunit of protein kinase A; implicated in mRNA nuclear export |
|
| YOR070C | 0.65 |
GYP1
|
Cis-golgi GTPase-activating protein (GAP) for the Rab family members Ypt1p (in vivo) and for Ypt1p, Sec4p, Ypt7p, and Ypt51p (in vitro); involved in vesicle docking and fusion |
|
| YHR095W | 0.65 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJL005W | 0.65 |
CYR1
|
Adenylate cyclase, required for cAMP production and cAMP-dependent protein kinase signaling; the cAMP pathway controls a variety of cellular processes, including metabolism, cell cycle, stress response, stationary phase, and sporulation |
|
| YBR208C | 0.65 |
DUR1,2
|
Urea amidolyase, contains both urea carboxylase and allophanate hydrolase activities, degrades urea to CO2 and NH3; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YOR027W | 0.64 |
STI1
|
Hsp90 cochaperone, interacts with the Ssa group of the cytosolic Hsp70 chaperones; activates the ATPase activity of Ssa1p; homolog of mammalian Hop protein |
|
| YAL028W | 0.64 |
FRT2
|
Tail-anchored endoplasmic reticulum membrane protein, interacts with homolog Frt1p but is not a substrate of calcineurin (unlike Frt1p), promotes growth in conditions of high Na+, alkaline pH, or cell wall stress; potential Cdc28p substrate |
|
| YMR090W | 0.63 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YDR313C | 0.61 |
PIB1
|
RING-type ubiquitin ligase of the endosomal and vacuolar membranes, binds phosphatidylinositol(3)-phosphate; contains a FYVE finger domain |
|
| YBL067C | 0.61 |
UBP13
|
Putative ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YHL025W | 0.61 |
SNF6
|
Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; functions interdependently in transcriptional activation with Snf2p and Snf5p |
|
| YAL066W | 0.60 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YAL063C | 0.60 |
FLO9
|
Lectin-like protein with similarity to Flo1p, thought to be expressed and involved in flocculation |
|
| YPR027C | 0.60 |
Putative protein of unknown function |
||
| YLR402W | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YKL177W | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene STE3 |
||
| YOR141C | 0.58 |
ARP8
|
Nuclear actin-related protein involved in chromatin remodeling, component of chromatin-remodeling enzyme complexes |
|
| YDR542W | 0.58 |
PAU10
|
Hypothetical protein |
|
| YGL059W | 0.58 |
PKP2
|
Mitochondrial protein kinase that negatively regulates activity of the pyruvate dehydrogenase complex by phosphorylating the ser-133 residue of the Pda1p subunit; acts in concert with kinase Pkp1p and phosphatases Ptc5p and Ptc6p |
|
| YKR096W | 0.56 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and cytoplasm; predicted to contain a PINc (PilT N terminus) domain |
||
| YMR107W | 0.56 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YLR403W | 0.56 |
SFP1
|
Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation |
|
| YMR279C | 0.56 |
Putative protein of unknown function; identified as a heat-induced gene in a high-throughout screen; YMR279C is not an essential gene |
||
| YOR215C | 0.55 |
AIM41
|
Putative protein of unknown function; the authentic protein is detected in highly purified mitochondria in high-throughput studies; null mutant displays altered rate of mitochondrial loss and reduced growth rate in minimal glycerol media |
|
| YAL047W-A | 0.55 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR278C | 0.55 |
Zinc-cluster protein; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; mutant shows moderate growth defect on caffeine; YLR278C is not an essential gene |
||
| YDR452W | 0.55 |
PPN1
|
Vacuolar endopolyphosphatase with a role in phosphate metabolism; functions as a homodimer |
|
| YPL147W | 0.55 |
PXA1
|
Subunit of a heterodimeric peroxisomal ATP-binding cassette transporter complex (Pxa1p-Pxa2p), required for import of long-chain fatty acids into peroxisomes; similarity to human adrenoleukodystrophy transporter and ALD-related proteins |
|
| YHR086W | 0.54 |
NAM8
|
RNA binding protein, component of the U1 snRNP protein; mutants are defective in meiotic recombination and in formation of viable spores, involved in the formation of DSBs through meiosis-specific splicing of MER2 pre-mRNA |
|
| YMR068W | 0.54 |
AVO2
|
Component of a complex containing the Tor2p kinase and other proteins, which may have a role in regulation of cell growth |
|
| YER075C | 0.54 |
PTP3
|
Phosphotyrosine-specific protein phosphatase involved in the inactivation of mitogen-activated protein kinase (MAPK) during osmolarity sensing; dephosporylates Hog1p MAPK and regulates its localization; localized to the cytoplasm |
Network of associatons between targets according to the STRING database.
First level regulatory network of GAT4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.7 | 4.0 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.6 | 5.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 2.1 | GO:0015976 | carbon utilization(GO:0015976) |
| 0.5 | 1.5 | GO:0015755 | fructose transport(GO:0015755) |
| 0.5 | 1.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.5 | 1.4 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.5 | 1.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.4 | 1.3 | GO:0035786 | protein complex oligomerization(GO:0035786) |
| 0.4 | 1.7 | GO:0036498 | mRNA splicing via endonucleolytic cleavage and ligation involved in unfolded protein response(GO:0030969) IRE1-mediated unfolded protein response(GO:0036498) mRNA splicing, via endonucleolytic cleavage and ligation(GO:0070054) |
| 0.4 | 1.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 1.0 | GO:0006343 | establishment of chromatin silencing(GO:0006343) |
| 0.3 | 0.3 | GO:0036257 | multivesicular body organization(GO:0036257) |
| 0.3 | 1.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 1.9 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.3 | 1.9 | GO:0060963 | positive regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0060963) |
| 0.3 | 2.2 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.3 | 1.5 | GO:0000173 | inactivation of MAPK activity involved in osmosensory signaling pathway(GO:0000173) |
| 0.3 | 0.3 | GO:0034225 | cellular response to zinc ion starvation(GO:0034224) regulation of transcription from RNA polymerase II promoter in response to zinc ion starvation(GO:0034225) |
| 0.3 | 1.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.3 | 1.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) thiamine diphosphate metabolic process(GO:0042357) |
| 0.3 | 0.3 | GO:0009118 | regulation of glycolytic process(GO:0006110) regulation of nucleoside metabolic process(GO:0009118) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) regulation of ATP metabolic process(GO:1903578) |
| 0.3 | 0.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 2.2 | GO:0015758 | glucose transport(GO:0015758) |
| 0.3 | 1.1 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.3 | 1.9 | GO:0019655 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.3 | 2.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 1.5 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.2 | 1.0 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.2 | 1.2 | GO:0032443 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.2 | 1.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.2 | 1.2 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 0.9 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.2 | 3.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.2 | 0.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.9 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.2 | 1.6 | GO:0051322 | anaphase(GO:0051322) |
| 0.2 | 1.1 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.2 | 0.7 | GO:0000709 | meiotic joint molecule formation(GO:0000709) |
| 0.2 | 0.6 | GO:0043419 | urea catabolic process(GO:0043419) |
| 0.2 | 2.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.2 | 0.6 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 4.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) |
| 0.2 | 1.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.6 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.2 | 0.2 | GO:0019627 | urea metabolic process(GO:0019627) |
| 0.2 | 1.6 | GO:0045912 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) negative regulation of gluconeogenesis(GO:0045721) negative regulation of carbohydrate metabolic process(GO:0045912) |
| 0.2 | 1.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 0.4 | GO:0090180 | regulation of vitamin metabolic process(GO:0030656) positive regulation of vitamin metabolic process(GO:0046136) regulation of thiamine biosynthetic process(GO:0070623) positive regulation of thiamine biosynthetic process(GO:0090180) |
| 0.2 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 1.2 | GO:0070542 | response to lipid(GO:0033993) response to oleic acid(GO:0034201) response to fatty acid(GO:0070542) cellular response to lipid(GO:0071396) cellular response to fatty acid(GO:0071398) cellular response to oleic acid(GO:0071400) |
| 0.2 | 0.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.2 | 0.5 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.2 | 0.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 0.6 | GO:0009190 | cAMP biosynthetic process(GO:0006171) cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.2 | 0.2 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 2.9 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.7 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.6 | GO:0061013 | regulation of mRNA catabolic process(GO:0061013) |
| 0.1 | 0.9 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.1 | 2.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.4 | GO:0006798 | polyphosphate catabolic process(GO:0006798) |
| 0.1 | 1.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.7 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.1 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0000348 | mRNA branch site recognition(GO:0000348) |
| 0.1 | 0.5 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 0.9 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose catabolic process(GO:0005987) |
| 0.1 | 0.3 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.1 | 0.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 0.2 | GO:0061647 | histone H3 acetylation(GO:0043966) histone H3-K9 acetylation(GO:0043970) histone H3-K14 acetylation(GO:0044154) histone H3-K9 modification(GO:0061647) |
| 0.1 | 1.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 2.0 | GO:0015918 | sterol transport(GO:0015918) |
| 0.1 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 1.1 | GO:0070272 | proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.3 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 3.7 | GO:0071940 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.1 | 1.2 | GO:0034087 | establishment of sister chromatid cohesion(GO:0034085) establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.1 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) histone demethylation(GO:0016577) demethylation(GO:0070988) |
| 0.1 | 0.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.1 | 2.7 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 0.4 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) copper ion homeostasis(GO:0055070) |
| 0.1 | 0.7 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.7 | GO:0071709 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.1 | 0.4 | GO:0000755 | cytogamy(GO:0000755) |
| 0.1 | 0.1 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.1 | 2.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.2 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 0.5 | GO:0031321 | ascospore-type prospore assembly(GO:0031321) |
| 0.1 | 0.2 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.1 | 0.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.1 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.1 | GO:0009847 | spore germination(GO:0009847) |
| 0.1 | 0.5 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 0.4 | GO:0034367 | DNA-templated transcriptional open complex formation(GO:0001112) protein-DNA complex remodeling(GO:0001120) macromolecular complex remodeling(GO:0034367) |
| 0.1 | 0.4 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of metal ion(GO:0051238) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.2 | GO:0098542 | immune effector process(GO:0002252) immune system process(GO:0002376) response to virus(GO:0009615) response to external biotic stimulus(GO:0043207) defense response to virus(GO:0051607) response to other organism(GO:0051707) defense response to other organism(GO:0098542) |
| 0.1 | 1.3 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 0.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.1 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 1.2 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.1 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 0.3 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.1 | 0.2 | GO:0000431 | regulation of transcription from RNA polymerase II promoter by galactose(GO:0000431) positive regulation of transcription from RNA polymerase II promoter by galactose(GO:0000435) |
| 0.1 | 0.3 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.4 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 1.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.5 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) |
| 0.1 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.2 | GO:0098630 | aggregation of unicellular organisms(GO:0098630) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0060194 | antisense RNA transcription(GO:0009300) regulation of antisense RNA transcription(GO:0060194) negative regulation of antisense RNA transcription(GO:0060195) |
| 0.0 | 0.9 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.4 | GO:0045996 | negative regulation of transcription by pheromones(GO:0045996) negative regulation of transcription from RNA polymerase II promoter by pheromones(GO:0046020) |
| 0.0 | 0.1 | GO:0019935 | cAMP-mediated signaling(GO:0019933) cyclic-nucleotide-mediated signaling(GO:0019935) |
| 0.0 | 0.2 | GO:0001080 | nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) nitrogen catabolite activation of transcription(GO:0090294) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0060237 | regulation of fungal-type cell wall organization(GO:0060237) |
| 0.0 | 0.0 | GO:0006952 | defense response(GO:0006952) |
| 0.0 | 0.7 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.2 | GO:0006113 | fermentation(GO:0006113) |
| 0.0 | 0.2 | GO:0006673 | inositolphosphoceramide metabolic process(GO:0006673) |
| 0.0 | 0.2 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.2 | GO:0097271 | protein localization to bud neck(GO:0097271) |
| 0.0 | 0.5 | GO:0000742 | karyogamy involved in conjugation with cellular fusion(GO:0000742) |
| 0.0 | 3.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0001308 | negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.1 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 0.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.1 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.4 | GO:0019400 | glycerol metabolic process(GO:0006071) alditol metabolic process(GO:0019400) |
| 0.0 | 0.5 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.3 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:1901977 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.4 | GO:0033617 | respiratory chain complex IV assembly(GO:0008535) mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 1.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0000902 | cell morphogenesis(GO:0000902) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.2 | GO:0019932 | second-messenger-mediated signaling(GO:0019932) |
| 0.0 | 0.3 | GO:0019878 | lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.2 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.2 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0072350 | tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 | 0.1 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) AMP metabolic process(GO:0046033) |
| 0.0 | 0.2 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.2 | GO:0001101 | response to acid chemical(GO:0001101) |
| 0.0 | 0.4 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0006077 | (1->6)-beta-D-glucan metabolic process(GO:0006077) |
| 0.0 | 0.0 | GO:0000087 | mitotic M phase(GO:0000087) M phase(GO:0000279) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.0 | GO:0034476 | U4 snRNA 3'-end processing(GO:0034475) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.0 | GO:0035822 | gene conversion(GO:0035822) |
| 0.0 | 0.0 | GO:0006784 | heme a biosynthetic process(GO:0006784) heme a metabolic process(GO:0046160) |
| 0.0 | 0.3 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.1 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.0 | GO:0001304 | progressive alteration of chromatin involved in replicative cell aging(GO:0001304) |
| 0.0 | 0.0 | GO:0042843 | D-xylose catabolic process(GO:0042843) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.7 | 2.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 1.5 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.3 | 0.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 2.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 1.0 | GO:0000817 | COMA complex(GO:0000817) |
| 0.2 | 3.2 | GO:0005619 | ascospore wall(GO:0005619) |
| 0.2 | 6.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 0.7 | GO:0045269 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.2 | 0.7 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.2 | 1.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.2 | 1.0 | GO:0034448 | EGO complex(GO:0034448) |
| 0.2 | 0.7 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.2 | 0.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 1.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 0.7 | GO:0045334 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle(GO:0030139) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 0.5 | GO:0000113 | nucleotide-excision repair factor 4 complex(GO:0000113) |
| 0.2 | 0.8 | GO:0072379 | ER membrane insertion complex(GO:0072379) TRC complex(GO:0072380) |
| 0.2 | 0.8 | GO:0032545 | CURI complex(GO:0032545) |
| 0.1 | 1.3 | GO:0045263 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.4 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.3 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 4.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.3 | GO:0031010 | Isw1 complex(GO:0016587) ISWI-type complex(GO:0031010) |
| 0.1 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0044233 | ERMES complex(GO:0032865) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:0071561 | organelle membrane contact site(GO:0044232) nucleus-vacuole junction(GO:0071561) |
| 0.1 | 1.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.9 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.1 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.1 | GO:0097002 | mitochondrial inner boundary membrane(GO:0097002) |
| 0.1 | 0.2 | GO:0030874 | nucleolar chromatin(GO:0030874) |
| 0.1 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.1 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0071013 | U2-type catalytic step 2 spliceosome(GO:0071007) catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0032221 | Rpd3S complex(GO:0032221) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0048188 | histone methyltransferase complex(GO:0035097) Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0070775 | NuA3 histone acetyltransferase complex(GO:0033100) H3 histone acetyltransferase complex(GO:0070775) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.4 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.1 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 7.1 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.1 | GO:0000328 | fungal-type vacuole lumen(GO:0000328) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0043332 | mating projection tip(GO:0043332) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0000131 | incipient cellular bud site(GO:0000131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 1.0 | 4.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.7 | 2.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 1.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.4 | 1.3 | GO:0008137 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 5.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.4 | 1.5 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.4 | 1.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.4 | 1.1 | GO:0015294 | solute:cation symporter activity(GO:0015294) |
| 0.3 | 1.0 | GO:0008972 | hydroxymethylpyrimidine kinase activity(GO:0008902) phosphomethylpyrimidine kinase activity(GO:0008972) |
| 0.3 | 1.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.3 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 0.7 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.3 | 1.3 | GO:0005186 | mating pheromone activity(GO:0000772) pheromone activity(GO:0005186) |
| 0.3 | 3.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.3 | 1.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.3 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 2.9 | GO:0099600 | transmembrane signaling receptor activity(GO:0004888) signaling receptor activity(GO:0038023) transmembrane receptor activity(GO:0099600) |
| 0.2 | 0.7 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.2 | 4.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 0.2 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 2.2 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.2 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.2 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 1.0 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.2 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 0.9 | GO:0097372 | NAD-dependent histone deacetylase activity (H3-K18 specific)(GO:0097372) |
| 0.2 | 0.7 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 2.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.2 | 0.8 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.3 | GO:0032452 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) histone demethylase activity(GO:0032452) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.1 | 0.4 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.7 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 1.1 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 1.0 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.9 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) signaling adaptor activity(GO:0035591) |
| 0.1 | 2.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 4.9 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.8 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 5.1 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.4 | GO:0016898 | lactate dehydrogenase activity(GO:0004457) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.1 | 0.3 | GO:0030414 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.1 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 2.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 1.0 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) mannose transmembrane transporter activity(GO:0015578) |
| 0.1 | 0.3 | GO:0052742 | phosphatidylinositol kinase activity(GO:0052742) |
| 0.1 | 3.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.3 | GO:0004026 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.1 | 0.2 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 0.2 | GO:0001156 | RNA polymerase III transcription factor binding(GO:0001025) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0019902 | phosphatase binding(GO:0019902) protein phosphatase binding(GO:0019903) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0022838 | substrate-specific channel activity(GO:0022838) |
| 0.0 | 0.1 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) |
| 0.0 | 0.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.2 | GO:0018456 | aryl-alcohol dehydrogenase (NAD+) activity(GO:0018456) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.5 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.1 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 1.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.0 | 0.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0008897 | holo-[acyl-carrier-protein] synthase activity(GO:0008897) |
| 0.0 | 2.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.0 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.0 | GO:0008143 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.0 | 0.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 1.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 32.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.5 | 4.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 0.7 | REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | Genes involved in The citric acid (TCA) cycle and respiratory electron transport |
| 0.2 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 0.3 | REACTOME TRIF MEDIATED TLR3 SIGNALING | Genes involved in TRIF mediated TLR3 signaling |
| 0.1 | 0.7 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.1 | 0.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.4 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.1 | 0.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.0 | 32.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |