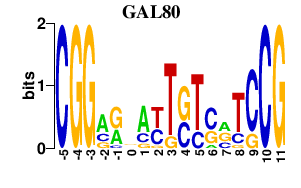
Results for GAL80
Z-value: 1.06
Transcription factors associated with GAL80
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GAL80
|
S000004515 | Transcriptional regulator involved in the repression of GAL genes |
Activity-expression correlation:
Activity profile of GAL80 motif
Sorted Z-values of GAL80 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YNL178W | 13.57 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YLR154W-B | 6.60 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-A | 6.57 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154C | 6.06 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YOL086C | 5.64 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YHR010W | 4.98 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YDL023C | 4.73 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YDR508C | 4.31 |
GNP1
|
High-affinity glutamine permease, also transports Leu, Ser, Thr, Cys, Met and Asn; expression is fully dependent on Grr1p and modulated by the Ssy1p-Ptr3p-Ssy5p (SPS) sensor of extracellular amino acids |
|
| YDR509W | 4.30 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YJR105W | 4.28 |
ADO1
|
Adenosine kinase, required for the utilization of S-adenosylmethionine (AdoMet); may be involved in recycling adenosine produced through the methyl cycle |
|
| YDR098C | 4.14 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YDL022W | 3.97 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YER102W | 3.93 |
RPS8B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps8Ap and has similarity to rat S8 ribosomal protein |
|
| YDL085C-A | 3.91 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YIL009W | 3.90 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YJL158C | 3.49 |
CIS3
|
Mannose-containing glycoprotein constituent of the cell wall; member of the PIR (proteins with internal repeats) family |
|
| YDL022C-A | 3.46 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene DIA3; identified by fungal homology and RT-PCR |
||
| YIL009C-A | 3.32 |
EST3
|
Component of the telomerase holoenzyme, involved in telomere replication |
|
| YPL112C | 3.18 |
PEX25
|
Peripheral peroxisomal membrane peroxin required for the regulation of peroxisome size and maintenance, recruits GTPase Rho1p to peroxisomes, induced by oleate, interacts with homologous protein Pex27p |
|
| YGR138C | 3.13 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YLR154W-C | 3.08 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YKR038C | 3.05 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YLR109W | 3.01 |
AHP1
|
Thiol-specific peroxiredoxin, reduces hydroperoxides to protect against oxidative damage; function in vivo requires covalent conjugation to Urm1p |
|
| YLR110C | 2.97 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YDR344C | 2.96 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR345C | 2.84 |
HXT3
|
Low affinity glucose transporter of the major facilitator superfamily, expression is induced in low or high glucose conditions |
|
| YFL022C | 2.76 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YDR279W | 2.74 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YMR009W | 2.74 |
ADI1
|
Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions |
|
| YEL056W | 2.71 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YML026C | 2.68 |
RPS18B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Ap and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YDR417C | 2.62 |
Hypothetical protein |
||
| YBR032W | 2.61 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YNL030W | 2.60 |
HHF2
|
One of two identical histone H4 proteins (see also HHF1); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity |
|
| YLR441C | 2.56 |
RPS1A
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Bp and has similarity to rat S3a ribosomal protein |
|
| YML063W | 2.54 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YOL085C | 2.50 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YKL182W | 2.43 |
FAS1
|
Beta subunit of fatty acid synthetase, which catalyzes the synthesis of long-chain saturated fatty acids; contains acetyltransacylase, dehydratase, enoyl reductase, malonyl transacylase, and palmitoyl transacylase activities |
|
| YMR199W | 2.41 |
CLN1
|
G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) |
|
| YOL039W | 2.40 |
RPP2A
|
Ribosomal protein P2 alpha, a component of the ribosomal stalk, which is involved in the interaction between translational elongation factors and the ribosome; regulates the accumulation of P1 (Rpp1Ap and Rpp1Bp) in the cytoplasm |
|
| YGR148C | 2.37 |
RPL24B
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Ap and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YNL327W | 2.36 |
EGT2
|
Glycosylphosphatidylinositol (GPI)-anchored cell wall endoglucanase required for proper cell separation after cytokinesis, expression is activated by Swi5p and tightly regulated in a cell cycle-dependent manner |
|
| YMR049C | 2.32 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YKR092C | 2.30 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YEL001C | 2.30 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YJR094W-A | 2.26 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YGR155W | 2.26 |
CYS4
|
Cystathionine beta-synthase, catalyzes the synthesis of cystathionine from serine and homocysteine, the first committed step in cysteine biosynthesis |
|
| YER001W | 2.25 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YBL092W | 2.24 |
RPL32
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L32 ribosomal protein; overexpression disrupts telomeric silencing |
|
| YIL052C | 2.20 |
RPL34B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Ap and has similarity to rat L34 ribosomal protein |
|
| YGR279C | 2.17 |
SCW4
|
Cell wall protein with similarity to glucanases; scw4 scw10 double mutants exhibit defects in mating |
|
| YMR194W | 2.14 |
RPL36A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, nearly identical to Rpl36Bp and has similarity to rat L36 ribosomal protein; binds to 5.8 S rRNA |
|
| YLR081W | 2.09 |
GAL2
|
Galactose permease, required for utilization of galactose; also able to transport glucose |
|
| YER074W | 2.09 |
RPS24A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Bp and has similarity to rat S24 ribosomal protein |
|
| YER023C-A | 2.08 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps verified gene PRO3; identified by gene-trapping, microarray expression analysis, and genome-wide homology searching |
||
| YMR083W | 2.04 |
ADH3
|
Mitochondrial alcohol dehydrogenase isozyme III; involved in the shuttling of mitochondrial NADH to the cytosol under anaerobic conditions and ethanol production |
|
| YLR355C | 2.04 |
ILV5
|
Acetohydroxyacid reductoisomerase, mitochondrial protein involved in branched-chain amino acid biosynthesis, also required for maintenance of wild-type mitochondrial DNA and found in mitochondrial nucleoids |
|
| YOL040C | 2.02 |
RPS15
|
Protein component of the small (40S) ribosomal subunit; has similarity to E. coli S19 and rat S15 ribosomal proteins |
|
| YMR032W | 2.00 |
HOF1
|
Bud neck-localized, SH3 domain-containing protein required for cytokinesis; regulates actomyosin ring dynamics and septin localization; interacts with the formins, Bni1p and Bnr1p, and with Cyk3p, Vrp1p, and Bni5p |
|
| YNR001W-A | 1.98 |
Dubious open reading frame unlikely to encode a functional protein; identified by homology |
||
| YGL039W | 1.94 |
Oxidoreductase, catalyzes NADPH-dependent reduction of the bicyclic diketone bicyclo[2.2.2]octane-2,6-dione (BCO2,6D) to the chiral ketoalcohol (1R,4S,6S)-6-hydroxybicyclo[2.2.2]octane-2-one (BCO2one6ol) |
||
| YER137C | 1.93 |
Putative protein of unknown function |
||
| YGR140W | 1.91 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YEL020C-B | 1.87 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene YEL020W-A; identified by fungal homology and RT-PCR |
||
| YNL289W | 1.87 |
PCL1
|
Pho85 cyclin of the Pcl1,2-like subfamily, involved in entry into the mitotic cell cycle and regulation of morphogenesis, localizes to sites of polarized cell growth |
|
| YPR063C | 1.83 |
ER-localized protein of unknown function |
||
| YEL054C | 1.79 |
RPL12A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Bp; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YLR108C | 1.78 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; YLR108C is not an esssential gene |
||
| YFR055W | 1.75 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YCL058C | 1.75 |
FYV5
|
Protein of unknown function, required for survival upon exposure to K1 killer toxin; involved in ion homeostasis |
|
| YPL014W | 1.72 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and to the nucleus |
||
| YJR145C | 1.65 |
RPS4A
|
Protein component of the small (40S) ribosomal subunit; mutation affects 20S pre-rRNA processing; identical to Rps4Bp and has similarity to rat S4 ribosomal protein |
|
| YGR139W | 1.65 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YER177W | 1.63 |
BMH1
|
14-3-3 protein, major isoform; controls proteome at post-transcriptional level, binds proteins and DNA, involved in regulation of many processes including exocytosis, vesicle transport, Ras/MAPK signaling, and rapamycin-sensitive signaling |
|
| YDL241W | 1.60 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YDL047W | 1.60 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YDR400W | 1.59 |
URH1
|
Uridine nucleosidase (uridine-cytidine N-ribohydrolase), cleaves N-glycosidic bonds in nucleosides; involved in the pyrimidine salvage and nicotinamide riboside salvage pathways |
|
| YIL123W | 1.58 |
SIM1
|
Protein of the SUN family (Sim1p, Uth1p, Nca3p, Sun4p) that may participate in DNA replication, promoter contains SCB regulation box at -300 bp indicating that expression may be cell cycle-regulated |
|
| YLR183C | 1.57 |
TOS4
|
Transcription factor that binds to a number of promoter regions, particularly promoters of some genes involved in pheromone response and cell cycle; potential Cdc28p substrate; expression is induced in G1 by bound SBF |
|
| YHR021C | 1.56 |
RPS27B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Ap and has similarity to rat S27 ribosomal protein |
|
| YER056C-A | 1.55 |
RPL34A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl34Bp and has similarity to rat L34 ribosomal protein |
|
| YGL040C | 1.53 |
HEM2
|
Delta-aminolevulinate dehydratase, a homo-octameric enzyme, catalyzes the conversion of delta-aminolevulinic acid to porphobilinogen, the second step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus |
|
| YCL064C | 1.49 |
CHA1
|
Catabolic L-serine (L-threonine) deaminase, catalyzes the degradation of both L-serine and L-threonine; required to use serine or threonine as the sole nitrogen source, transcriptionally induced by serine and threonine |
|
| YLR351C | 1.49 |
NIT3
|
Nit protein, one of two proteins in S. cerevisiae with similarity to the Nit domain of NitFhit from fly and worm and to the mouse and human Nit protein which interacts with the Fhit tumor suppressor; nitrilase superfamily member |
|
| YCL063W | 1.49 |
VAC17
|
Protein involved in vacuole inheritance; acts as a vacuole-specific receptor for myosin Myo2p |
|
| YIL158W | 1.48 |
AIM20
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YLR344W | 1.48 |
RPL26A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Bp and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YMR144W | 1.47 |
Putative protein of unknown function; localized to the nucleus; YMR144W is not an essential gene |
||
| YBR112C | 1.47 |
CYC8
|
General transcriptional co-repressor, acts together with Tup1p; also acts as part of a transcriptional co-activator complex that recruits the SWI/SNF and SAGA complexes to promoters |
|
| YIL056W | 1.43 |
VHR1
|
Transcriptional activator, required for the vitamin H-responsive element (VHRE) mediated induction of VHT1 (Vitamin H transporter) and BIO5 (biotin biosynthesis intermediate transporter) in response to low biotin concentrations |
|
| YGL034C | 1.43 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL075W | 1.42 |
RPL31A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Bp and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YFR056C | 1.41 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YDR156W | 1.41 |
RPA14
|
RNA polymerase I subunit A14 |
|
| YNL031C | 1.39 |
HHT2
|
One of two identical histone H3 proteins (see also HHT1); core histone required for chromatin assembly, involved in heterochromatin-mediated telomeric and HM silencing; regulated by acetylation, methylation, and mitotic phosphorylation |
|
| YHR094C | 1.38 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YKL120W | 1.36 |
OAC1
|
Mitochondrial inner membrane transporter, transports oxaloacetate, sulfate, and thiosulfate; member of the mitochondrial carrier family |
|
| YLR185W | 1.35 |
RPL37A
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl37Bp and to rat L37 ribosomal protein |
|
| YML103C | 1.32 |
NUP188
|
Subunit of the nuclear pore complex (NPC), involved in the structural organization of the complex and of the nuclear envelope, also involved in nuclear envelope permeability, interacts with Pom152p and Nic96p |
|
| YJL196C | 1.31 |
ELO1
|
Elongase I, medium-chain acyl elongase, catalyzes carboxy-terminal elongation of unsaturated C12-C16 fatty acyl-CoAs to C16-C18 fatty acids |
|
| YMR105W-A | 1.31 |
Putative protein of unknown function |
||
| YGR251W | 1.30 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YOR092W | 1.29 |
ECM3
|
Non-essential protein of unknown function; involved in signal transduction and the genotoxic response; induced rapidly in response to treatment with 8-methoxypsoralen and UVA irradiation |
|
| YHR216W | 1.28 |
IMD2
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, expression is induced by mycophenolic acid resulting in resistance to the drug, expression is repressed by nutrient limitation |
|
| YOR130C | 1.27 |
ORT1
|
Ornithine transporter of the mitochondrial inner membrane, exports ornithine from mitochondria as part of arginine biosynthesis; human ortholog is associated with hyperammonaemia-hyperornithinaemia-homocitrullinuria (HHH) syndrome |
|
| YLR257W | 1.26 |
Putative protein of unknown function |
||
| YEL038W | 1.26 |
UTR4
|
Protein of unknown function with sequence similarity to 2,3-diketo-5-methylthiopentyl-1-phosphate enolase-phosphatases, found in both the cytoplasm and nucleus |
|
| YBR113W | 1.25 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene CYC8 |
||
| YPL265W | 1.24 |
DIP5
|
Dicarboxylic amino acid permease, mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate; also a transporter for Gln, Asn, Ser, Ala, and Gly |
|
| YML123C | 1.24 |
PHO84
|
High-affinity inorganic phosphate (Pi) transporter and low-affinity manganese transporter; regulated by Pho4p and Spt7p; mutation confers resistance to arsenate; exit from the ER during maturation requires Pho86p |
|
| YER088C-A | 1.24 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL009C | 1.23 |
LEU1
|
Isopropylmalate isomerase, catalyzes the second step in the leucine biosynthesis pathway |
|
| YNL058C | 1.21 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole; YNL058C is not an essential gene |
||
| YJL216C | 1.21 |
Protein of unknown function, similar to alpha-D-glucosidases; transcriptionally activated by both Pdr8p and Yrm1p, along with transporters and other genes involved in the pleiotropic drug resistance (PDR) phenomenon |
||
| YDR033W | 1.20 |
MRH1
|
Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p |
|
| YDR112W | 1.20 |
IRC2
|
Dubious open reading frame, unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps YDR111C; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YHR043C | 1.20 |
DOG2
|
2-deoxyglucose-6-phosphate phosphatase, member of a family of low molecular weight phosphatases, similar to Dog1p, induced by oxidative and osmotic stress, confers 2-deoxyglucose resistance when overexpressed |
|
| YLR042C | 1.19 |
Protein of unknown function; localizes to the cytoplasm; YLL042C is not an essential gene |
||
| YOR096W | 1.16 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YER028C | 1.15 |
MIG3
|
Probable transcriptional repressor involved in response to toxic agents such as hydroxyurea that inhibit ribonucleotide reductase; phosphorylation by Snf1p or the Mec1p pathway inactivates Mig3p, allowing induction of damage response genes |
|
| YIL148W | 1.14 |
RPL40A
|
Fusion protein, identical to Rpl40Bp, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YCL018W | 1.14 |
LEU2
|
Beta-isopropylmalate dehydrogenase (IMDH), catalyzes the third step in the leucine biosynthesis pathway |
|
| YNL057W | 1.11 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YFR031C-A | 1.11 |
RPL2A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Bp and has similarity to E. coli L2 and rat L8 ribosomal proteins |
|
| YLR437C | 1.10 |
Putative protein of unknown function; epitope tagged protein localizes to the cytoplasm |
||
| YGR090W | 1.08 |
UTP22
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YAL038W | 1.07 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YDL137W | 1.05 |
ARF2
|
ADP-ribosylation factor, GTPase of the Ras superfamily involved in regulation of coated formation vesicles in intracellular trafficking within the Golgi; functionally interchangeable with Arf1p |
|
| YGR040W | 1.02 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YIL053W | 1.01 |
RHR2
|
Constitutively expressed isoform of DL-glycerol-3-phosphatase; involved in glycerol biosynthesis, induced in response to both anaerobic and, along with the Hor2p/Gpp2p isoform, osmotic stress |
|
| YMR246W | 1.01 |
FAA4
|
Long chain fatty acyl-CoA synthetase, regulates protein modification during growth in the presence of ethanol, functions to incorporate palmitic acid into phospholipids and neutral lipids |
|
| YGL106W | 1.00 |
MLC1
|
Essential light chain for Myo1p, light chain for Myo2p; stabilizes Myo2p by binding to the neck region; interacts with Myo1p, Iqg1p, and Myo2p to coordinate formation and contraction of the actomyosin ring with targeted membrane deposition |
|
| YDR111C | 0.98 |
ALT2
|
Putative alanine transaminase (glutamic pyruvic transaminase) |
|
| YDR395W | 0.97 |
SXM1
|
Nuclear transport factor (karyopherin) involved in protein transport between the cytoplasm and nucleoplasm; similar to Nmd5p, Cse1p, Lph2p, and the human cellular apoptosis susceptibility protein, CAS1 |
|
| YGR055W | 0.97 |
MUP1
|
High affinity methionine permease, integral membrane protein with 13 putative membrane-spanning regions; also involved in cysteine uptake |
|
| YDL211C | 0.96 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YML111W | 0.95 |
BUL2
|
Component of the Rsp5p E3-ubiquitin ligase complex, involved in intracellular amino acid permease sorting, functions in heat shock element mediated gene expression, essential for growth in stress conditions, functional homolog of BUL1 |
|
| YLL045C | 0.95 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YJL200C | 0.92 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YKL218C | 0.91 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YCR102W-A | 0.90 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOR119C | 0.89 |
RIO1
|
Essential serine kinase involved in cell cycle progression and processing of the 20S pre-rRNA into mature 18S rRNA |
|
| YGL201C | 0.89 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YKL110C | 0.89 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YML064C | 0.88 |
TEM1
|
GTP-binding protein of the ras superfamily involved in termination of M-phase; controls actomyosin and septin dynamics during cytokinesis |
|
| YHL003C | 0.88 |
LAG1
|
Ceramide synthase component, involved in synthesis of ceramide from C26(acyl)-coenzyme A and dihydrosphingosine or phytosphingosine, functionally equivalent to Lac1p |
|
| YHR140W | 0.88 |
Putative integral membrane protein of unknown function |
||
| YLR328W | 0.87 |
NMA1
|
Nicotinic acid mononucleotide adenylyltransferase, involved in pathways of NAD biosynthesis, including the de novo, NAD(+) salvage, and nicotinamide riboside salvage pathways |
|
| YOR342C | 0.87 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YPL250W-A | 0.87 |
Identified by fungal homology and RT-PCR |
||
| YDR037W | 0.87 |
KRS1
|
Lysyl-tRNA synthetase; also identified as a negative regulator of general control of amino acid biosynthesis |
|
| YAR029W | 0.87 |
Member of DUP240 gene family but contains no transmembrane domains; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YKR093W | 0.87 |
PTR2
|
Integral membrane peptide transporter, mediates transport of di- and tri-peptides; conserved protein that contains 12 transmembrane domains; PTR2 expression is regulated by the N-end rule pathway via repression by Cup9p |
|
| YPR074C | 0.86 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YLR411W | 0.85 |
CTR3
|
High-affinity copper transporter of the plasma membrane, acts as a trimer; gene is disrupted by a Ty2 transposon insertion in many laboratory strains of S. cerevisiae |
|
| YKL030W | 0.85 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; partially overlaps the verified gene MAE1 |
||
| YFL026W | 0.85 |
STE2
|
Receptor for alpha-factor pheromone; seven transmembrane-domain GPCR that interacts with both pheromone and a heterotrimeric G protein to initiate the signaling response that leads to mating between haploid a and alpha cells |
|
| YPR014C | 0.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPR014C is not an essential gene |
||
| YPR170W-B | 0.83 |
Putative protein of unknown function, conserved in fungi; partially overlaps the dubious genes YPR169W-A, YPR170W-A and YRP170C |
||
| YBR118W | 0.82 |
TEF2
|
Translational elongation factor EF-1 alpha; also encoded by TEF1; functions in the binding reaction of aminoacyl-tRNA (AA-tRNA) to ribosomes |
|
| YBR106W | 0.81 |
PHO88
|
Probable membrane protein, involved in phosphate transport; pho88 pho86 double null mutant exhibits enhanced synthesis of repressible acid phosphatase at high inorganic phosphate concentrations |
|
| YGL031C | 0.81 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YLL027W | 0.81 |
ISA1
|
Mitochondrial matrix protein involved in biogenesis of the iron-sulfur (Fe/S) cluster of Fe/S proteins, isa1 deletion causes loss of mitochondrial DNA and respiratory deficiency; depletion reduces growth on nonfermentable carbon sources |
|
| YML102W | 0.80 |
CAC2
|
Component of the chromatin assembly complex (with Rlf2p and Msi1p) that assembles newly synthesized histones onto recently replicated DNA, required for building functional kinetochores, conserved from yeast to humans |
|
| YCR102C | 0.78 |
Putative protein of unknown function; involved in copper metabolism; similar to C.carbonum toxD gene; YCR102C is not an essential gene |
||
| YMR008C | 0.78 |
PLB1
|
Phospholipase B (lysophospholipase) involved in lipid metabolism, required for deacylation of phosphatidylcholine and phosphatidylethanolamine but not phosphatidylinositol |
|
| YNR016C | 0.78 |
ACC1
|
Acetyl-CoA carboxylase, biotin containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA; required for de novo biosynthesis of long-chain fatty acids |
|
| YJR114W | 0.77 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSM7/YJR113C |
||
| YGL030W | 0.77 |
RPL30
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L30 ribosomal protein; involved in pre-rRNA processing in the nucleolus; autoregulates splicing of its transcript |
|
| YMR290C | 0.76 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YLR043C | 0.76 |
TRX1
|
Cytoplasmic thioredoxin isoenzyme of the thioredoxin system which protects cells against oxidative and reductive stress, forms LMA1 complex with Pbi2p, acts as a cofactor for Tsa1p, required for ER-Golgi transport and vacuole inheritance |
|
| YJL198W | 0.75 |
PHO90
|
Low-affinity phosphate transporter; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YOL155C | 0.75 |
HPF1
|
Haze-protective mannoprotein that reduces the particle size of aggregated proteins in white wines |
|
| YDR041W | 0.74 |
RSM10
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S10 ribosomal protein; essential for viability, unlike most other mitoribosomal proteins |
|
| YOR029W | 0.74 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YHR208W | 0.74 |
BAT1
|
Mitochondrial branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during logarithmic phase and repressed during stationary phase |
|
| YDR510W | 0.73 |
SMT3
|
Ubiquitin-like protein of the SUMO family, conjugated to lysine residues of target proteins; regulates chromatid cohesion, chromosome segregation, APC-mediated proteolysis, DNA replication and septin ring dynamics |
|
| YPL211W | 0.73 |
NIP7
|
Nucleolar protein required for 60S ribosome subunit biogenesis, constituent of 66S pre-ribosomal particles; physically interacts with Nop8p and the exosome subunit Rrp43p |
|
| YJR123W | 0.72 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YMR290W-A | 0.71 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YKR075C | 0.71 |
Protein of unknown function; similar to YOR062Cp and Reg1p; expression regulated by glucose and Rgt1p; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YJR104C | 0.71 |
SOD1
|
Cytosolic superoxide dismutase; some mutations are analogous to those that cause ALS (amyotrophic lateral sclerosis) in humans |
|
| YIL163C | 0.71 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YHR162W | 0.70 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the mitochondrion |
||
| YCL050C | 0.69 |
APA1
|
Diadenosine 5',5''-P1,P4-tetraphosphate phosphorylase I (AP4A phosphorylase), involved in catabolism of bis(5'-nucleosidyl) tetraphosphates; has similarity to Apa2p |
|
| YDR040C | 0.69 |
ENA1
|
P-type ATPase sodium pump, involved in Na+ and Li+ efflux to allow salt tolerance |
|
| YIR031C | 0.69 |
DAL7
|
Malate synthase, role in allantoin degradation unknown; expression sensitive to nitrogen catabolite repression and induced by allophanate, an intermediate in allantoin degradation |
|
| YDR047W | 0.69 |
HEM12
|
Uroporphyrinogen decarboxylase, catalyzes the fifth step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus; activity inhibited by Cu2+, Zn2+, Fe2+, Fe3+ and sulfhydryl-specific reagents |
|
| YIL164C | 0.68 |
NIT1
|
Nitrilase, member of the nitrilase branch of the nitrilase superfamily; in closely related species and other S. cerevisiae strain backgrounds YIL164C and adjacent ORF, YIL165C, likely constitute a single ORF encoding a nitrilase gene |
|
| YDL205C | 0.67 |
HEM3
|
Phorphobilinogen deaminase, catalyzes the conversion of 4-porphobilinogen to hydroxymethylbilane, the third step in the heme biosynthetic pathway; localizes to both the cytoplasm and nucleus; expression is regulated by Hap2p-Hap3p |
|
| YGL148W | 0.67 |
ARO2
|
Bifunctional chorismate synthase and flavin reductase, catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate (EPSP) to form chorismate, which is a precursor to aromatic amino acids |
|
| YNL239W | 0.66 |
LAP3
|
Cysteine aminopeptidase with homocysteine-thiolactonase activity; protects cells against homocysteine toxicity; has bleomycin hydrolase activity in vitro; transcription is regulated by galactose via Gal4p; orthologous to human BLMH |
|
| YPL143W | 0.66 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YGL105W | 0.66 |
ARC1
|
Protein that binds tRNA and methionyl- and glutamyl-tRNA synthetases (Mes1p and Gus1p), delivering tRNA to them, stimulating catalysis, and ensuring their localization to the cytoplasm; also binds quadruplex nucleic acids |
|
| YDR008C | 0.66 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDR155C | 0.64 |
CPR1
|
Cytoplasmic peptidyl-prolyl cis-trans isomerase (cyclophilin), catalyzes the cis-trans isomerization of peptide bonds N-terminal to proline residues; binds the drug cyclosporin A |
|
| YOR028C | 0.64 |
CIN5
|
Basic leucine zipper transcriptional factor of the yAP-1 family that mediates pleiotropic drug resistance and salt tolerance; localizes constitutively to the nucleus |
|
| YJR113C | 0.64 |
RSM7
|
Mitochondrial ribosomal protein of the small subunit, has similarity to E. coli S7 ribosomal protein |
|
| YNL069C | 0.63 |
RPL16B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Ap, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YBR084C-A | 0.62 |
RPL19A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Bp and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
Network of associatons between targets according to the STRING database.
First level regulatory network of GAL80
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 1.3 | 14.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.3 | 8.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 1.2 | 5.9 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 1.0 | 4.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.9 | 2.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.8 | 3.4 | GO:0000296 | spermine transport(GO:0000296) |
| 0.8 | 7.4 | GO:0000947 | amino acid catabolic process to alcohol via Ehrlich pathway(GO:0000947) |
| 0.8 | 2.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.7 | 3.0 | GO:0098742 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.7 | 2.1 | GO:2000877 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.6 | 2.3 | GO:0019346 | transsulfuration(GO:0019346) |
| 0.5 | 1.6 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 1.6 | GO:0042454 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) ribonucleoside catabolic process(GO:0042454) cytidine metabolic process(GO:0046087) |
| 0.5 | 4.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.5 | 2.0 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.5 | 1.5 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.5 | 2.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.5 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) C4-dicarboxylate transport(GO:0015740) |
| 0.4 | 1.3 | GO:0046039 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 0.4 | 2.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.4 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.4 | 1.9 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.4 | 57.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 2.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.3 | 2.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 1.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.3 | 1.7 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.3 | 1.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 2.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.3 | 1.2 | GO:0071478 | response to UV(GO:0009411) cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.3 | 2.4 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.3 | 4.7 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.2 | 2.6 | GO:0090529 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.2 | 0.7 | GO:0009164 | nucleoside catabolic process(GO:0009164) glycosyl compound catabolic process(GO:1901658) |
| 0.2 | 0.7 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.2 | 0.9 | GO:1900434 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.2 | 0.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.9 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin structure organization(GO:0031032) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 1.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.9 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.2 | 1.7 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.2 | 0.6 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.2 | 5.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 1.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.2 | 0.5 | GO:0001196 | regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001196) negative regulation of mating-type specific transcription from RNA polymerase II promoter(GO:0001198) negative regulation of cellular amine metabolic process(GO:0033239) regulation of cellular amine catabolic process(GO:0033241) negative regulation of mating-type specific transcription, DNA-templated(GO:0045894) positive regulation of mating-type specific transcription, DNA-templated(GO:0045895) |
| 0.2 | 0.5 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.2 | 0.5 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.2 | 0.8 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 0.7 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.2 | 0.5 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 0.6 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 1.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.9 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.1 | 0.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.6 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.8 | GO:0009102 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 0.8 | GO:0048255 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.1 | 0.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) |
| 0.1 | 0.5 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.1 | 0.6 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.1 | 1.0 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.1 | 0.9 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.8 | GO:0030031 | cell projection organization(GO:0030030) cell projection assembly(GO:0030031) |
| 0.1 | 1.3 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.1 | 0.4 | GO:0000092 | mitotic anaphase B(GO:0000092) plasmid partitioning(GO:0030541) |
| 0.1 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.0 | GO:0031070 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.1 | 2.8 | GO:0008361 | regulation of cell size(GO:0008361) |
| 0.1 | 0.2 | GO:0009090 | homoserine biosynthetic process(GO:0009090) |
| 0.1 | 0.3 | GO:0009097 | isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.5 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 1.5 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.1 | 0.3 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.1 | 0.8 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 1.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 1.9 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0038032 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.6 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 1.0 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.3 | GO:0050810 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.1 | 3.6 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 1.6 | GO:0032005 | pheromone-dependent signal transduction involved in conjugation with cellular fusion(GO:0000750) signal transduction involved in conjugation with cellular fusion(GO:0032005) |
| 0.1 | 0.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 0.2 | GO:0048209 | regulation of COPII vesicle coating(GO:0003400) regulation of vesicle targeting, to, from or within Golgi(GO:0048209) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.3 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 0.5 | GO:0052803 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.1 | 0.3 | GO:0090158 | regulation of phosphatidylinositol dephosphorylation(GO:0060304) endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 1.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0051457 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0032365 | intracellular lipid transport(GO:0032365) |
| 0.0 | 2.8 | GO:0000747 | conjugation with cellular fusion(GO:0000747) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.0 | 3.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0042435 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 0.0 | 1.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 3.6 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.0 | 0.3 | GO:0031120 | tRNA pseudouridine synthesis(GO:0031119) snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.6 | GO:0051666 | actin cortical patch localization(GO:0051666) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.3 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.0 | 1.1 | GO:0006694 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0044070 | regulation of anion transport(GO:0044070) |
| 0.0 | 0.3 | GO:0030846 | termination of RNA polymerase II transcription, poly(A)-coupled(GO:0030846) |
| 0.0 | 0.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.7 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:0015883 | FAD transport(GO:0015883) |
| 0.0 | 0.2 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.0 | 0.3 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.9 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
| 0.0 | 0.2 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0070096 | mitochondrial outer membrane translocase complex assembly(GO:0070096) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.1 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.0 | 0.2 | GO:0051668 | localization within membrane(GO:0051668) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 2.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0034401 | chromatin organization involved in regulation of transcription(GO:0034401) |
| 0.0 | 0.2 | GO:0030474 | spindle pole body duplication(GO:0030474) |
| 0.0 | 0.2 | GO:0044396 | actin cortical patch assembly(GO:0000147) actin cortical patch organization(GO:0044396) |
| 0.0 | 0.5 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:0055075 | cellular potassium ion homeostasis(GO:0030007) potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.7 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.0 | GO:0019419 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.0 | 0.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.3 | 4.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.1 | 13.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.8 | 2.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.8 | 2.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.6 | 3.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.5 | 2.0 | GO:0044697 | HICS complex(GO:0044697) |
| 0.5 | 1.9 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.5 | 3.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.5 | 1.8 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 35.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.4 | 23.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 4.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 1.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 3.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 0.7 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.2 | 1.0 | GO:0032545 | CURI complex(GO:0032545) |
| 0.2 | 0.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0008275 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 0.1 | 1.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.0 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 3.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.7 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 1.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0043529 | GET complex(GO:0043529) |
| 0.1 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.6 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.1 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 6.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 0.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.7 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 0.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.3 | GO:0044445 | cytosolic part(GO:0044445) |
| 0.1 | 0.6 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.3 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.5 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 4.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.0 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.3 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.6 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.3 | GO:0099512 | supramolecular fiber(GO:0099512) polymeric cytoskeletal fiber(GO:0099513) |
| 0.0 | 0.8 | GO:0005815 | microtubule organizing center(GO:0005815) spindle pole body(GO:0005816) |
| 0.0 | 0.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.0 | 0.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 2.2 | 8.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.3 | 3.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.2 | 7.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 1.1 | 4.4 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.9 | 2.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.8 | 3.4 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.6 | 2.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.6 | 2.4 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.6 | 2.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 1.2 | GO:0015645 | long-chain fatty acid-CoA ligase activity(GO:0004467) fatty acid ligase activity(GO:0015645) |
| 0.6 | 3.5 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.5 | 1.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.5 | 2.6 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.5 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.5 | 1.4 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.4 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.4 | 1.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 1.6 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.4 | 2.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 1.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 0.7 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 1.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 3.8 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.3 | 2.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.3 | 60.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 0.8 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.3 | 0.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.3 | 1.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 6.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.3 | 4.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.2 | 3.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.5 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.2 | 1.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 0.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 0.8 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.2 | 5.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 0.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 3.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 0.6 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.1 | 1.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 0.4 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 1.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.5 | GO:0016725 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 4.1 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.1 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 2.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 1.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.3 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.1 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.6 | GO:0008144 | drug binding(GO:0008144) |
| 0.1 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.7 | GO:0015079 | potassium ion transmembrane transporter activity(GO:0015079) |
| 0.1 | 0.2 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 0.2 | GO:0008251 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.7 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 0.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.2 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 2.1 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0072509 | divalent inorganic cation transmembrane transporter activity(GO:0072509) |
| 0.0 | 0.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 0.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 16.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 1.0 | 4.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.9 | 2.8 | REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | Genes involved in Insulin receptor signalling cascade |
| 0.9 | 2.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.7 | 12.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.4 | 1.3 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.3 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 0.7 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.2 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.0 | 0.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |