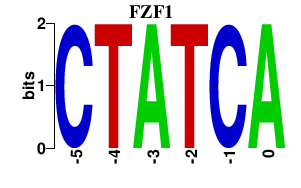
Results for FZF1
Z-value: 0.63
Transcription factors associated with FZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FZF1
|
S000003223 | Transcription factor involved in sulfite metabolism |
Activity-expression correlation:
Activity profile of FZF1 motif
Sorted Z-values of FZF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YER137C | 1.73 |
Putative protein of unknown function |
||
| YLR154W-A | 1.61 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YDR098C | 1.60 |
GRX3
|
Hydroperoxide and superoxide-radical responsive glutathione-dependent oxidoreductase; monothiol glutaredoxin subfamily member along with Grx4p and Grx5p; protects cells from oxidative damage |
|
| YLR154W-B | 1.54 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154C | 1.37 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YMR049C | 1.27 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YGR040W | 1.27 |
KSS1
|
Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response; the KSS1 gene is nonfunctional in S288C strains and functional in W303 strains |
|
| YER097W | 1.26 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YLR154W-C | 1.18 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YAL033W | 1.14 |
POP5
|
Subunit of both RNase MRP, which cleaves pre-rRNA, and nuclear RNase P, which cleaves tRNA precursors to generate mature 5' ends |
|
| YGR039W | 1.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YOR028C | 1.03 |
CIN5
|
Basic leucine zipper transcriptional factor of the yAP-1 family that mediates pleiotropic drug resistance and salt tolerance; localizes constitutively to the nucleus |
|
| YKR038C | 0.92 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YIL009W | 0.92 |
FAA3
|
Long chain fatty acyl-CoA synthetase, has a preference for C16 and C18 fatty acids; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YJL200C | 0.88 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YKR092C | 0.88 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YOL086C | 0.80 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YDL211C | 0.79 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the vacuole |
||
| YKL218C | 0.79 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YLR257W | 0.75 |
Putative protein of unknown function |
||
| YMR290W-A | 0.72 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YGL209W | 0.72 |
MIG2
|
Protein containing zinc fingers, involved in repression, along with Mig1p, of SUC2 (invertase) expression by high levels of glucose; binds to Mig1p-binding sites in SUC2 promoter |
|
| YOR107W | 0.71 |
RGS2
|
Negative regulator of glucose-induced cAMP signaling; directly activates the GTPase activity of the heterotrimeric G protein alpha subunit Gpa2p |
|
| YJL011C | 0.71 |
RPC17
|
RNA polymerase III subunit C17; physically interacts with C31, C11, and TFIIIB70; may be involved in the recruitment of pol III by the preinitiation complex |
|
| YDR002W | 0.70 |
YRB1
|
Ran GTPase binding protein; involved in nuclear protein import and RNA export, ubiquitin-mediated protein degradation during the cell cycle; shuttles between the nucleus and cytoplasm; is essential; homolog of human RanBP1 |
|
| YJL162C | 0.70 |
JJJ2
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YCR018C | 0.69 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YDR279W | 0.66 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YJL112W | 0.66 |
MDV1
|
Peripheral protein of the cytosolic face of the mitochondrial outer membrane, required for mitochondrial fission; interacts with Fis1p and with the dynamin-related GTPase Dnm1p; contains WD repeats |
|
| YHR181W | 0.64 |
SVP26
|
Integral membrane protein of the early Golgi apparatus and endoplasmic reticulum, involved in COP II vesicle transport; may also function to promote retention of proteins in the early Golgi compartment |
|
| YGR293C | 0.62 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YGR294W |
||
| YIL009C-A | 0.61 |
EST3
|
Component of the telomerase holoenzyme, involved in telomere replication |
|
| YHR094C | 0.59 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YJL198W | 0.58 |
PHO90
|
Low-affinity phosphate transporter; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YDR502C | 0.58 |
SAM2
|
S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) |
|
| YGR294W | 0.57 |
PAU12
|
Hypothetical protein |
|
| YFR056C | 0.57 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YPL062W | 0.56 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YIL074C | 0.55 |
SER33
|
3-phosphoglycerate dehydrogenase, catalyzes the first step in serine and glycine biosynthesis; isozyme of Ser3p |
|
| YLR449W | 0.54 |
FPR4
|
Peptidyl-prolyl cis-trans isomerase (PPIase) (proline isomerase) localized to the nucleus; catalyzes isomerization of proline residues in histones H3 and H4, which affects lysine methylation of those histones |
|
| YOR030W | 0.54 |
DFG16
|
Probable multiple transmembrane protein, involved in diploid invasive and pseudohyphal growth upon nitrogen starvation; required for accumulation of processed Rim101p |
|
| YGL201C | 0.54 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YCR019W | 0.54 |
MAK32
|
Protein necessary for structural stability of L-A double-stranded RNA-containing particles |
|
| YMR119W | 0.54 |
ASI1
|
Putative integral membrane E3 ubiquitin ligase; acts with Asi2p and Asi3p to ensure the fidelity of SPS-sensor signalling by maintaining the dormant repressed state of gene expression in the absence of inducing signals |
|
| YFR055W | 0.52 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YLR344W | 0.51 |
RPL26A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Bp and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YKL110C | 0.50 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YLR411W | 0.49 |
CTR3
|
High-affinity copper transporter of the plasma membrane, acts as a trimer; gene is disrupted by a Ty2 transposon insertion in many laboratory strains of S. cerevisiae |
|
| YEL029C | 0.48 |
BUD16
|
Putative pyridoxal kinase, key enzyme in vitamin B6 metabolism; involved in maintaining levels of pyridoxal 5'-phosphate, the active form of vitamin B6; required for genome integrity; homolog of E. coli PdxK; involved in bud-site selection |
|
| YOL103W | 0.48 |
ITR2
|
Myo-inositol transporter with strong similarity to the major myo-inositol transporter Itr1p, member of the sugar transporter superfamily; expressed constitutively |
|
| YLR333C | 0.47 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YOR342C | 0.46 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YMR136W | 0.45 |
GAT2
|
Protein containing GATA family zinc finger motifs; similar to Gln3p and Dal80p; expression repressed by leucine |
|
| YMR290C | 0.42 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YDL055C | 0.42 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YGR181W | 0.41 |
TIM13
|
Mitochondrial intermembrane space protein, forms a complex with TIm8p that mediates import and insertion of a subset of polytopic inner membrane proteins; may prevent aggregation of incoming proteins in a chaperone-like manner |
|
| YER159C | 0.40 |
BUR6
|
Subunit of a heterodimeric NC2 transcription regulator complex with Ncb2p; complex binds to TBP and can repress transcription by preventing preinitiation complex assembly or stimulate activated transcription; homologous to human NC2alpha |
|
| YPL128C | 0.40 |
TBF1
|
Telobox-containing general regulatory factor; binds to TTAGGG repeats within subtelomeric anti-silencing regions (STARs) and possibly throughout the genome and mediates their insulating capacity by blocking silent chromatin propagation |
|
| YDL241W | 0.39 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YER098W | 0.38 |
UBP9
|
Ubiquitin carboxyl-terminal hydrolase, ubiquitin-specific protease that cleaves ubiquitin-protein fusions |
|
| YMR135W-A | 0.38 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBL063W | 0.37 |
KIP1
|
Kinesin-related motor protein required for mitotic spindle assembly and chromosome segregation; functionally redundant with Cin8p |
|
| YKL122C | 0.37 |
SRP21
|
Subunit of the signal recognition particle (SRP), which functions in protein targeting to the endoplasmic reticulum membrane; not found in mammalian SRP; forms a pre-SRP structure in the nucleolus that is translocated to the cytoplasm |
|
| YEL053W-A | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YEL054C |
||
| YBR023C | 0.36 |
CHS3
|
Chitin synthase III, catalyzes the transfer of N-acetylglucosamine (GlcNAc) to chitin; required for synthesis of the majority of cell wall chitin, the chitin ring during bud emergence, and spore wall chitosan |
|
| YBR044C | 0.36 |
TCM62
|
Protein involved in the assembly of the mitochondrial succinate dehydrogenase complex; putative chaperone |
|
| YPL177C | 0.36 |
CUP9
|
Homeodomain-containing transcriptional repressor of PTR2, which encodes a major peptide transporter; imported peptides activate ubiquitin-dependent proteolysis, resulting in degradation of Cup9p and de-repression of PTR2 transcription |
|
| YDR339C | 0.35 |
FCF1
|
Essential nucleolar protein that is a component of the SSU (small subunit) processome involved in the pre-rRNA processing steps of 40S ribosomal subunit biogenesis; contains a PINc domain; copurifies with Faf1p |
|
| YHR180W-A | 0.34 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps dubious ORF YHR180C-B and long terminal repeat YHRCsigma3 |
||
| YER001W | 0.34 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YDR089W | 0.34 |
Protein of unknown function; deletion confers resistance to Nickel |
||
| YML009C | 0.34 |
MRPL39
|
Mitochondrial ribosomal protein of the large subunit |
|
| YBR126W-A | 0.34 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YPL055C | 0.34 |
LGE1
|
Protein of unknown function; null mutant forms abnormally large cells, and homozygous diploid null mutant displays delayed premeiotic DNA synthesis and reduced efficiency of meiotic nuclear division |
|
| YKL183C-A | 0.34 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YDL048C | 0.34 |
STP4
|
Protein containing a Kruppel-type zinc-finger domain; has similarity to Stp1p, Stp2p, and Stp3p |
|
| YLR425W | 0.34 |
TUS1
|
Guanine nucleotide exchange factor (GEF) that functions to modulate Rho1p activity as part of the cell integrity signaling pathway; multicopy suppressor of tor2 mutation and ypk1 ypk2 double mutation; potential Cdc28p substrate |
|
| YNL302C | 0.34 |
RPS19B
|
Protein component of the small (40S) ribosomal subunit, required for assembly and maturation of pre-40 S particles; mutations in human RPS19 are associated with Diamond Blackfan anemia; nearly identical to Rps19Ap |
|
| YLR407W | 0.33 |
Putative protein of unknown function; YLR407W is not an essential gene |
||
| YJR094C | 0.33 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YHR140W | 0.33 |
Putative integral membrane protein of unknown function |
||
| YKR093W | 0.33 |
PTR2
|
Integral membrane peptide transporter, mediates transport of di- and tri-peptides; conserved protein that contains 12 transmembrane domains; PTR2 expression is regulated by the N-end rule pathway via repression by Cup9p |
|
| YJL168C | 0.33 |
SET2
|
Histone methyltransferase with a role in transcriptional elongation, methylates a lysine residue of histone H3; associates with the C-terminal domain of Rpo21p; histone methylation activity is regulated by phosphorylation status of Rpo21p |
|
| YKL165C | 0.32 |
MCD4
|
Protein involved in glycosylphosphatidylinositol (GPI) anchor synthesis; multimembrane-spanning protein that localizes to the endoplasmic reticulum; highly conserved among eukaryotes |
|
| YHR005C-A | 0.32 |
MRS11
|
Essential protein of the mitochondrial intermembrane space, forms a complex with Tim9p (TIM10 complex) that mediates insertion of hydrophobic proteins at the inner membrane, has homology to Mrs5p, which is also involved in this process |
|
| YER131W | 0.32 |
RPS26B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps26Ap and has similarity to rat S26 ribosomal protein |
|
| YER012W | 0.32 |
PRE1
|
Beta 4 subunit of the 20S proteasome; localizes to the nucleus throughout the cell cycle |
|
| YNR054C | 0.32 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YMR106C | 0.32 |
YKU80
|
Subunit of the telomeric Ku complex (Yku70p-Yku80p), involved in telomere length maintenance, structure and telomere position effect; relocates to sites of double-strand cleavage to promote nonhomologous end joining during DSB repair |
|
| YMR098C | 0.32 |
ATP25
|
Mitochondrial protein required for the stability of Oli1p (Atp9p) mRNA and for the Oli1p ring formation; null mutant displays decreased frequency of mitochondrial genome loss (petite formation) |
|
| YNL119W | 0.32 |
NCS2
|
Protein with a role in urmylation and in invasive and pseudohyphal growth; inhibits replication of Brome mosaic virus in S. cerevisiae, which is a model system for studying replication of positive-strand RNA viruses in their natural hosts |
|
| YLR455W | 0.32 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
||
| YDR025W | 0.31 |
RPS11A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Bp and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YLR143W | 0.31 |
Putative protein of unknown function; green fluorescent protein (GFP)-tagged protein localizes to the cytoplasm; YLR143W is not an essential gene |
||
| YDL047W | 0.31 |
SIT4
|
Type 2A-related serine-threonine phosphatase that functions in the G1/S transition of the mitotic cycle; cytoplasmic and nuclear protein that modulates functions mediated by Pkc1p including cell wall and actin cytoskeleton organization |
|
| YGR027C | 0.31 |
RPS25A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Bp and has similarity to rat S25 ribosomal protein |
|
| YPL178W | 0.31 |
CBC2
|
Small subunit of the heterodimeric cap binding complex that also contains Sto1p, component of the spliceosomal commitment complex; interacts with Npl3p, possibly to package mRNA for export from the nucleus; contains an RNA-binding motif |
|
| YJR094W-A | 0.30 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YBR244W | 0.30 |
GPX2
|
Phospholipid hydroperoxide glutathione peroxidase induced by glucose starvation that protects cells from phospholipid hydroperoxides and nonphospholipid peroxides during oxidative stress |
|
| YOR344C | 0.30 |
TYE7
|
Serine-rich protein that contains a basic-helix-loop-helix (bHLH) DNA binding motif; binds E-boxes of glycolytic genes and contributes to their activation; may function as a transcriptional activator in Ty1-mediated gene expression |
|
| YPR104C | 0.30 |
FHL1
|
Transcriptional activator with similarity to DNA-binding domain of Drosophila forkhead but unable to bind DNA in vitro; required for rRNA processing; isolated as a suppressor of splicing factor prp4 |
|
| YOR130C | 0.29 |
ORT1
|
Ornithine transporter of the mitochondrial inner membrane, exports ornithine from mitochondria as part of arginine biosynthesis; human ortholog is associated with hyperammonaemia-hyperornithinaemia-homocitrullinuria (HHH) syndrome |
|
| YBR143C | 0.29 |
SUP45
|
Polypeptide release factor involved in translation termination; mutant form acts as a recessive omnipotent suppressor |
|
| YNL155W | 0.28 |
Putative protein of unknown function, contains DHHC domain, also predicted to have thiol-disulfide oxidoreductase active site |
||
| YBR300C | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YBR301W; YBR300C is not an essential gene |
||
| YHR208W | 0.28 |
BAT1
|
Mitochondrial branched-chain amino acid aminotransferase, homolog of murine ECA39; highly expressed during logarithmic phase and repressed during stationary phase |
|
| YGR195W | 0.28 |
SKI6
|
3'-to-5' phosphorolytic exoribonuclease that is a subunit of the exosome; required for 3' processing of the 5.8S rRNA; involved in 3' to 5' mRNA degradation and translation inhibition of non-poly(A) mRNAs |
|
| YNL120C | 0.28 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; deletion enhances replication of Brome mosaic virus in S. cerevisiae, but likely due to effects on the overlapping gene |
||
| YER016W | 0.28 |
BIM1
|
Microtubule-binding protein that together with Kar9p makes up the cortical microtubule capture site and delays the exit from mitosis when the spindle is oriented abnormally |
|
| YIL008W | 0.28 |
URM1
|
Ubiquitin-like protein with weak sequence similarity to ubiquitin; depends on the E1-like activating enzyme Uba4p; molecular function of the Urm1p pathway is unknown, but it is required for normal growth, particularly at high temperature |
|
| YOR008C-A | 0.27 |
Putative protein of unknown function, includes a potential transmembrane domain; deletion results in slightly lengthened telomeres |
||
| YBL085W | 0.27 |
BOI1
|
Protein implicated in polar growth, functionally redundant with Boi2p; interacts with bud-emergence protein Bem1p; contains an SH3 (src homology 3) domain and a PH (pleckstrin homology) domain |
|
| YDL192W | 0.27 |
ARF1
|
ADP-ribosylation factor, GTPase of the Ras superfamily involved in regulation of coated vesicle formation in intracellular trafficking within the Golgi; functionally interchangeable with Arf2p |
|
| YER183C | 0.26 |
FAU1
|
5,10-methenyltetrahydrofolate synthetase, involved in folic acid biosynthesis |
|
| YCL022C | 0.26 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps verified gene KCC4/YCL024W |
||
| YKR026C | 0.26 |
GCN3
|
Alpha subunit of the translation initiation factor eIF2B, the guanine-nucleotide exchange factor for eIF2; activity subsequently regulated by phosphorylated eIF2; first identified as a positive regulator of GCN4 expression |
|
| YPR118W | 0.26 |
MRI1
|
Methylthioribose-1-phosphate isomerase, catalyzes the isomerization of 5-methylthioribose-1-phosphate to 5-methylthioribulose-1-phosphate in the methionine salvage pathway |
|
| YNL154C | 0.26 |
YCK2
|
Palmitoylated, plasma membrane-bound casein kinase I isoform; shares redundant functions with Yck1p in morphogenesis, proper septin assembly, endocytic trafficking; provides an essential function overlapping with that of Yck1p |
|
| YEL007W | 0.26 |
Putative protein with sequence similarity to S. pombe gti1+ (gluconate transport inducer 1) |
||
| YBR210W | 0.25 |
ERV15
|
Protein involved in export of proteins from the endoplasmic reticulum, has similarity to Erv14p |
|
| YAR018C | 0.25 |
KIN3
|
Nonessential protein kinase with unknown cellular role |
|
| YDR266C | 0.25 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments;green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; contains a RING finger domain |
||
| YHR212W-A | 0.25 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YCR099C | 0.25 |
Putative protein of unknown function |
||
| YBL092W | 0.25 |
RPL32
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L32 ribosomal protein; overexpression disrupts telomeric silencing |
|
| YPR119W | 0.24 |
CLB2
|
B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome |
|
| YER147C-A | 0.24 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR005C | 0.24 |
MAF1
|
Negative regulator of RNA polymerase III; component of several signaling pathways that repress polymerase III transcription in response to changes in cellular environment; targets the initiation factor TFIIIB |
|
| YNL032W | 0.24 |
SIW14
|
Tyrosine phosphatase that plays a role in actin filament organization and endocytosis; localized to the cytoplasm |
|
| YNR061C | 0.24 |
Putative protein of unknown function |
||
| YNL016W | 0.24 |
PUB1
|
Poly(A)+ RNA-binding protein, abundant mRNP-component protein that binds mRNA and is required for stability of a number of mRNAs; not reported to associate with polyribosomes |
|
| YCR006C | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL159C-A | 0.24 |
Putative protein of unknown function; identified by sequence comparison with hemiascomycetous yeast species |
||
| YER130C | 0.24 |
Hypothetical protein |
||
| YBR243C | 0.24 |
ALG7
|
UDP-N-acetyl-glucosamine-1-P transferase, transfers Glc-Nac-P from UDP-GlcNac to Dol-P in the ER in the first step of the dolichol pathway of protein asparagine-linked glycosylation; inhibited by tunicamycin |
|
| YPL265W | 0.23 |
DIP5
|
Dicarboxylic amino acid permease, mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate; also a transporter for Gln, Asn, Ser, Ala, and Gly |
|
| YMR039C | 0.23 |
SUB1
|
Transcriptional coactivator, facilitates elongation by influencing enzymes that modify RNAP II, acts in a peroxide resistance pathway involving Rad2p; suppressor of TFIIB mutations |
|
| YBR249C | 0.23 |
ARO4
|
3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, catalyzes the first step in aromatic amino acid biosynthesis and is feedback-inhibited by tyrosine or high concentrations of phenylalanine or tryptophan |
|
| YKL189W | 0.23 |
HYM1
|
Component of the RAM signaling network that is involved in regulation of Ace2p activity and cellular morphogenesis, interacts with Kic1p and Sog2p, localizes to sites of polarized growth during budding and during the mating response |
|
| YLL025W | 0.23 |
PAU17
|
Putative protein of unknown function; YLL025W is not an essential gene |
|
| YGR228W | 0.23 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF SMI1/YGR229C |
||
| YKL042W | 0.23 |
SPC42
|
Central plaque component of spindle pole body (SPB); involved in SPB duplication, may facilitate attachment of the SPB to the nuclear membrane |
|
| YNL298W | 0.22 |
CLA4
|
Cdc42p activated signal transducing kinase of the PAK (p21-activated kinase) family, involved in septin ring assembly and cytokinesis; directly phosphorylates septins Cdc3p and Cdc10p; other yeast PAK family members are Ste20p and Skm1p |
|
| YER054C | 0.22 |
GIP2
|
Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism; contains a conserved motif (GVNK motif) that is also found in Gac1p, Pig1p, and Pig2p |
|
| YOR043W | 0.22 |
WHI2
|
Protein required, with binding partner Psr1p, for full activation of the general stress response, possibly through Msn2p dephosphorylation; regulates growth during the diauxic shift; negative regulator of G1 cyclin expression |
|
| YCR097W-A | 0.22 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; identified by homology to a hemiascomycetous yeast protein |
||
| YOR345C | 0.22 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YKL001C | 0.22 |
MET14
|
Adenylylsulfate kinase, required for sulfate assimilation and involved in methionine metabolism |
|
| YIL050W | 0.22 |
PCL7
|
Pho85p cyclin of the Pho80p subfamily, forms a functional kinase complex with Pho85p which phosphorylates Mmr1p and is regulated by Pho81p; involved in glycogen metabolism, expression is cell-cycle regulated |
|
| YJR010W | 0.22 |
MET3
|
ATP sulfurylase, catalyzes the primary step of intracellular sulfate activation, essential for assimilatory reduction of sulfate to sulfide, involved in methionine metabolism |
|
| YLR048W | 0.22 |
RPS0B
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Ap; required for maturation of 18S rRNA along with Rps0Ap; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YPL143W | 0.22 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YMR177W | 0.22 |
MMT1
|
Putative metal transporter involved in mitochondrial iron accumulation; closely related to Mmt2p |
|
| YOR047C | 0.22 |
STD1
|
Protein involved in control of glucose-regulated gene expression; interacts with protein kinase Snf1p, glucose sensors Snf3p and Rgt2p, and TATA-binding protein Spt15p; acts as a regulator of the transcription factor Rgt1p |
|
| YLR025W | 0.22 |
SNF7
|
One of four subunits of the endosomal sorting complex required for transport III (ESCRT-III); involved in the sorting of transmembrane proteins into the multivesicular body (MVB) pathway; recruited from the cytoplasm to endosomal membranes |
|
| YOR315W | 0.21 |
SFG1
|
Nuclear protein, putative transcription factor required for growth of superficial pseudohyphae (which do not invade the agar substrate) but not for invasive pseudohyphal growth; may act together with Phd1p; potential Cdc28p substrate |
|
| YEL072W | 0.21 |
RMD6
|
Protein required for sporulation |
|
| YGR264C | 0.21 |
MES1
|
Methionyl-tRNA synthetase, forms a complex with glutamyl-tRNA synthetase (Gus1p) and Arc1p, which increases the catalytic efficiency of both tRNA synthetases; also has a role in nuclear export of tRNAs |
|
| YCL007C | 0.21 |
Dubious ORF unlikely to encode a protein; overlaps verified ORF YCL005W-A; mutations in YCL007C were thought to confer sensitivity to calcofluor white, but this phenotype was later shown to be due to the defect in YCL005W-A |
||
| YIL051C | 0.21 |
MMF1
|
Mitochondrial protein involved in maintenance of the mitochondrial genome |
|
| YFR028C | 0.21 |
CDC14
|
Protein phosphatase required for mitotic exit; located in the nucleolus until liberated by the FEAR and Mitotic Exit Network in anaphase, enabling it to act on key substrates to effect a decrease in CDK/B-cyclin activity and mitotic exit |
|
| YOR326W | 0.21 |
MYO2
|
One of two type V myosin motors (along with MYO4) involved in actin-based transport of cargos; required for the polarized delivery of secretory vesicles, the vacuole, late Golgi elements, peroxisomes, and the mitotic spindle |
|
| YDR088C | 0.21 |
SLU7
|
RNA splicing factor, required for ATP-independent portion of 2nd catalytic step of spliceosomal RNA splicing; interacts with Prp18p; contains zinc knuckle domain |
|
| YOL160W | 0.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR191W | 0.21 |
PEX13
|
Integral peroxisomal membrane required for the translocation of peroxisomal matrix proteins, interacts with the PTS1 signal recognition factor Pex5p and the PTS2 signal recognition factor Pex7p, forms a complex with Pex14p and Pex17p |
|
| YHR010W | 0.21 |
RPL27A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl27Bp and has similarity to rat L27 ribosomal protein |
|
| YKL156W | 0.20 |
RPS27A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps27Bp and has similarity to rat S27 ribosomal protein |
|
| YBR202W | 0.20 |
MCM7
|
Component of the hexameric MCM complex, which is important for priming origins of DNA replication in G1 and becomes an active ATP-dependent helicase that promotes DNA melting and elongation when activated by Cdc7p-Dbf4p in S-phase |
|
| YOR324C | 0.20 |
FRT1
|
Tail-anchored endoplasmic reticulum membrane protein that is a substrate of the phosphatase calcineurin, interacts with homolog Frt2p, promotes cell growth in conditions of high Na+, alkaline pH, and cell wall stress |
|
| YGR188C | 0.20 |
BUB1
|
Protein kinase that forms a complex with Mad1p and Bub3p that is crucial in the checkpoint mechanism required to prevent cell cycle progression into anaphase in the presence of spindle damage, associates with centromere DNA via Skp1p |
|
| YGR155W | 0.20 |
CYS4
|
Cystathionine beta-synthase, catalyzes the synthesis of cystathionine from serine and homocysteine, the first committed step in cysteine biosynthesis |
|
| YOL085C | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOL085W-A |
||
| YNL064C | 0.20 |
YDJ1
|
Protein chaperone involved in regulation of the HSP90 and HSP70 functions; involved in protein translocation across membranes; member of the DnaJ family |
|
| YGL123C-A | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene RPS2/YGL123W |
||
| YOR381W-A | 0.20 |
Putative protein of unknown function; identified by fungal homology and RT-PCR |
||
| YLR406C | 0.20 |
RPL31B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Ap and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YOR104W | 0.20 |
PIN2
|
Protein that induces appearance of [PIN+] prion when overproduced |
|
| YGR265W | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF MES1/YGR264C, which encodes methionyl-tRNA synthetase |
||
| YGL123W | 0.19 |
RPS2
|
Protein component of the small (40S) subunit, essential for control of translational accuracy; has similarity to E. coli S5 and rat S2 ribosomal proteins |
|
| YDR043C | 0.19 |
NRG1
|
Transcriptional repressor that recruits the Cyc8p-Tup1p complex to promoters; mediates glucose repression and negatively regulates a variety of processes including filamentous growth and alkaline pH response |
|
| YML006C | 0.19 |
GIS4
|
CAAX box containing protein of unknown function, proposed to be involved in the RAS/cAMP signaling pathway |
|
| YPL112C | 0.19 |
PEX25
|
Peripheral peroxisomal membrane peroxin required for the regulation of peroxisome size and maintenance, recruits GTPase Rho1p to peroxisomes, induced by oleate, interacts with homologous protein Pex27p |
|
| YBR247C | 0.19 |
ENP1
|
Protein associated with U3 and U14 snoRNAs, required for pre-rRNA processing and 40S ribosomal subunit synthesis; localized in the nucleus and concentrated in the nucleolus |
|
| YDR477W | 0.19 |
SNF1
|
AMP-activated serine/threonine protein kinase found in a complex containing Snf4p and members of the Sip1p/Sip2p/Gal83p family; required for transcription of glucose-repressed genes, thermotolerance, sporulation, and peroxisome biogenesis |
|
| YJL122W | 0.19 |
ALB1
|
Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit; interacts directly with Arx1p; responsible for Tif6p recycling defects in absence of Rei1p |
|
| YGL117W | 0.19 |
Putative protein of unknown function |
||
| YOR029W | 0.19 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YOL012C | 0.19 |
HTZ1
|
Histone variant H2AZ, exchanged for histone H2A in nucleosomes by the SWR1 complex; involved in transcriptional regulation through prevention of the spread of silent heterochromatin |
|
| YKR074W | 0.19 |
AIM29
|
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YFR020W | 0.19 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YIL004C | 0.19 |
BET1
|
Type II membrane protein required for vesicular transport between the endoplasmic reticulum and Golgi complex; v-SNARE with similarity to synaptobrevins |
|
| YPR187W | 0.19 |
RPO26
|
RNA polymerase subunit ABC23, common to RNA polymerases I, II, and III; part of central core; similar to bacterial omega subunit |
|
| YNL075W | 0.18 |
IMP4
|
Component of the SSU processome, which is required for pre-18S rRNA processing; interacts with Mpp10p; member of a superfamily of proteins that contain a sigma(70)-like motif and associate with RNAs |
|
| YEL001C | 0.18 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YEL050C | 0.18 |
RML2
|
Mitochondrial ribosomal protein of the large subunit, has similarity to E. coli L2 ribosomal protein; fat21 mutant allele causes inability to utilize oleate and may interfere with activity of the Adr1p transcription factor |
|
| YJL223C | 0.18 |
PAU1
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YDR395W | 0.18 |
SXM1
|
Nuclear transport factor (karyopherin) involved in protein transport between the cytoplasm and nucleoplasm; similar to Nmd5p, Cse1p, Lph2p, and the human cellular apoptosis susceptibility protein, CAS1 |
|
| YFL017W-A | 0.18 |
SMX2
|
Core Sm protein Sm G; part of heteroheptameric complex (with Smb1p, Smd1p, Smd2p, Smd3p, Sme1p, and Smx3p) that is part of the spliceosomal U1, U2, U4, and U5 snRNPs; homolog of human Sm G |
|
| YML063W | 0.18 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YOR226C | 0.18 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
Network of associatons between targets according to the STRING database.
First level regulatory network of FZF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.1 | GO:0036170 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.2 | 1.2 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.2 | 0.7 | GO:0038032 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 0.9 | GO:0031057 | negative regulation of histone modification(GO:0031057) |
| 0.2 | 0.8 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.2 | 1.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.2 | 0.9 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.2 | 0.5 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.4 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 0.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 1.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.4 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 1.1 | GO:0034965 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.1 | 0.4 | GO:0030541 | plasmid partitioning(GO:0030541) |
| 0.1 | 0.5 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.1 | 0.3 | GO:0035955 | regulation of oligopeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035950) negative regulation of oligopeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035952) regulation of dipeptide transport by regulation of transcription from RNA polymerase II promoter(GO:0035953) negative regulation of dipeptide transport by negative regulation of transcription from RNA polymerase II promoter(GO:0035955) negative regulation of oligopeptide transport(GO:2000877) negative regulation of dipeptide transport(GO:2000879) |
| 0.1 | 1.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 0.6 | GO:0007535 | donor selection(GO:0007535) |
| 0.1 | 0.9 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.4 | GO:0001015 | snoRNA transcription from an RNA polymerase II promoter(GO:0001015) negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 0.3 | GO:1900544 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.3 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.1 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) negative regulation of homeostatic process(GO:0032845) |
| 0.1 | 0.3 | GO:0070637 | nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.2 | GO:0070814 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.3 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.1 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.6 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.1 | 0.5 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.5 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.1 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.1 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) |
| 0.1 | 0.2 | GO:0009097 | isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.5 | GO:0070873 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.1 | 0.5 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.2 | GO:0006798 | polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.1 | 1.0 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.1 | GO:0001324 | age-dependent general metabolic decline involved in chronological cell aging(GO:0001323) age-dependent response to oxidative stress involved in chronological cell aging(GO:0001324) |
| 0.1 | 1.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.2 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.2 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 0.1 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.3 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.1 | 0.1 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.3 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.1 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) |
| 0.1 | 0.2 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.1 | 0.1 | GO:0000092 | mitotic anaphase B(GO:0000092) |
| 0.1 | 0.5 | GO:0034221 | cell wall chitin biosynthetic process(GO:0006038) fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.1 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.3 | GO:0072668 | obsolete tubulin complex biogenesis(GO:0072668) |
| 0.1 | 0.2 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.0 | 0.6 | GO:0007118 | budding cell apical bud growth(GO:0007118) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.0 | GO:1903339 | inactivation of MAPK activity involved in cell wall organization or biogenesis(GO:0000200) regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903137) negative regulation of MAPK cascade involved in cell wall organization or biogenesis(GO:1903138) negative regulation of cell wall organization or biogenesis(GO:1903339) |
| 0.0 | 0.1 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 5.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:1903338 | regulation of cell wall organization or biogenesis(GO:1903338) |
| 0.0 | 0.1 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.0 | GO:1903322 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.0 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) regulation of cellular amine catabolic process(GO:0033241) |
| 0.0 | 0.1 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) intracellular copper ion transport(GO:0015680) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 2.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0019660 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 0.0 | 0.1 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.2 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.0 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0009088 | threonine metabolic process(GO:0006566) threonine biosynthetic process(GO:0009088) |
| 0.0 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.1 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) barbed-end actin filament capping(GO:0051016) actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.2 | GO:0042559 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0035753 | maintenance of DNA trinucleotide repeats(GO:0035753) |
| 0.0 | 0.0 | GO:1901654 | response to ketone(GO:1901654) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0070481 | nuclear-transcribed mRNA catabolic process, non-stop decay(GO:0070481) |
| 0.0 | 0.2 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.3 | GO:0009304 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:2000002 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 2.1 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.0 | 0.1 | GO:0032886 | regulation of microtubule-based process(GO:0032886) regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.5 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0015883 | FAD transport(GO:0015883) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 1.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.0 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.1 | GO:0016072 | rRNA metabolic process(GO:0016072) rRNA catabolic process(GO:0016075) ncRNA catabolic process(GO:0034661) |
| 0.0 | 0.0 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0043007 | maintenance of rDNA(GO:0043007) |
| 0.0 | 0.0 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.7 | GO:0007124 | pseudohyphal growth(GO:0007124) |
| 0.0 | 0.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.0 | 0.1 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.0 | GO:0009071 | serine family amino acid catabolic process(GO:0009071) |
| 0.0 | 0.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.1 | GO:0006721 | terpenoid metabolic process(GO:0006721) terpenoid biosynthetic process(GO:0016114) farnesyl diphosphate biosynthetic process(GO:0045337) farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.1 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) macromolecular complex remodeling(GO:0034367) |
| 0.0 | 0.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.0 | 0.0 | GO:0007534 | gene conversion at mating-type locus(GO:0007534) |
| 0.0 | 0.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:0032545 | CURI complex(GO:0032545) |
| 0.1 | 0.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.1 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 3.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0044284 | mitochondrial crista junction(GO:0044284) |
| 0.1 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0030869 | RENT complex(GO:0030869) |
| 0.1 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0005823 | central plaque of spindle pole body(GO:0005823) |
| 0.0 | 0.3 | GO:0005724 | nuclear heterochromatin(GO:0005720) nuclear telomeric heterochromatin(GO:0005724) telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 0.2 | GO:0070772 | PAS complex(GO:0070772) |
| 0.0 | 0.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 0.0 | 1.4 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 3.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0097344 | Rix1 complex(GO:0097344) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.0 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.1 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.0 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.6 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0031499 | TRAMP complex(GO:0031499) |
| 0.0 | 0.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0033180 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.5 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.1 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.0 | 0.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0000111 | nucleotide-excision repair factor 2 complex(GO:0000111) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 0.6 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.2 | 0.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.2 | 0.5 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.2 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 1.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 1.1 | GO:0004526 | ribonuclease MRP activity(GO:0000171) ribonuclease P activity(GO:0004526) |
| 0.1 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.3 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.1 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.1 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 7.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.0 | 0.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0019888 | phosphatase regulator activity(GO:0019208) protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0008186 | RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0000156 | phosphorelay response regulator activity(GO:0000156) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) RNA polymerase binding(GO:0070063) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.0 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.3 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 1.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.0 | 0.1 | GO:0042123 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.1 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.0 | 0.0 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0015248 | oxysterol binding(GO:0008142) sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.1 | REACTOME MITOTIC M M G1 PHASES | Genes involved in Mitotic M-M/G1 phases |
| 0.0 | 0.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |