Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
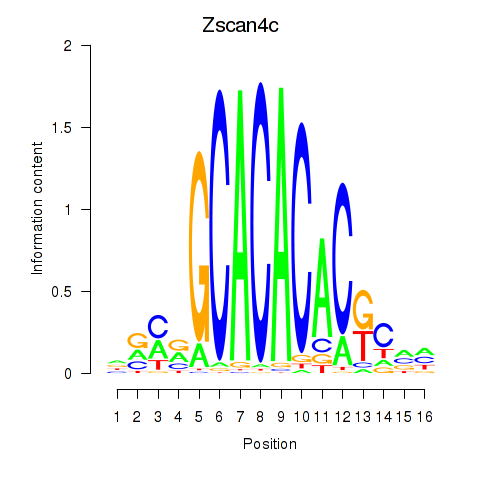
Results for Zscan4c
Z-value: 0.61
Transcription factors associated with Zscan4c
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zscan4f | rn6_v1_chr7_-_17859653_17859653 | 0.11 | 4.3e-02 | Click! |
Activity profile of Zscan4c motif
Sorted Z-values of Zscan4c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44746549 | 26.75 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr7_-_140483693 | 22.19 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chr10_+_27973681 | 18.04 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chr5_-_62621737 | 13.91 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr10_-_107539658 | 13.73 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr10_-_107539465 | 12.30 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr1_-_8751198 | 11.57 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr13_+_101181994 | 11.07 |
ENSRNOT00000052407
|
Susd4
|
sushi domain containing 4 |
| chr6_+_92229686 | 10.72 |
ENSRNOT00000046085
|
Atl1
|
atlastin GTPase 1 |
| chr2_-_154418920 | 10.27 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chr2_-_154418629 | 9.94 |
ENSRNOT00000076274
ENSRNOT00000076165 |
Plch1
|
phospholipase C, eta 1 |
| chr9_+_16862248 | 9.85 |
ENSRNOT00000080104
ENSRNOT00000024824 |
Ttbk1
|
tau tubulin kinase 1 |
| chr4_-_180234804 | 9.79 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr18_+_59748444 | 9.40 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr15_+_108908607 | 9.24 |
ENSRNOT00000089455
|
Zic2
|
Zic family member 2 |
| chr1_+_170262156 | 9.22 |
ENSRNOT00000024077
ENSRNOT00000085395 |
Cckbr
|
cholecystokinin B receptor |
| chr17_-_9791781 | 9.02 |
ENSRNOT00000090536
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr18_-_14016713 | 9.01 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr2_+_239415046 | 8.44 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr1_-_239057732 | 7.36 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr8_-_62987182 | 7.29 |
ENSRNOT00000070885
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr5_-_7941822 | 7.17 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr2_+_236625357 | 6.81 |
ENSRNOT00000081248
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr12_+_31934343 | 6.61 |
ENSRNOT00000011181
|
Tmem132d
|
transmembrane protein 132D |
| chr17_-_9792007 | 6.05 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr4_-_727691 | 6.04 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr1_+_87248489 | 5.76 |
ENSRNOT00000028091
|
Dpf1
|
double PHD fingers 1 |
| chr1_+_247084228 | 5.66 |
ENSRNOT00000020475
|
Plpp6
|
phospholipid phosphatase 6 |
| chr13_+_82369493 | 5.53 |
ENSRNOT00000003733
|
Sell
|
selectin L |
| chr4_-_114853868 | 5.49 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr15_-_52399074 | 5.08 |
ENSRNOT00000018440
|
Xpo7
|
exportin 7 |
| chrX_+_135348436 | 4.68 |
ENSRNOT00000008868
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr1_-_265798167 | 4.47 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr19_+_51317425 | 4.36 |
ENSRNOT00000019298
|
Cdh13
|
cadherin 13 |
| chr20_+_42966140 | 4.19 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr17_-_77687456 | 3.94 |
ENSRNOT00000045765
ENSRNOT00000081645 |
Frmd4a
|
FERM domain containing 4A |
| chr5_+_133864798 | 3.68 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chrX_+_70438617 | 3.33 |
ENSRNOT00000076671
|
Arr3
|
arrestin 3 |
| chr13_-_90022269 | 3.09 |
ENSRNOT00000035498
|
Ly9
|
lymphocyte antigen 9 |
| chr3_+_138974871 | 2.98 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr20_+_48335540 | 2.95 |
ENSRNOT00000000352
|
Cd24
|
CD24 molecule |
| chr3_-_107760550 | 2.63 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr9_+_94879745 | 2.21 |
ENSRNOT00000080482
|
Atg16l1
|
autophagy related 16-like 1 |
| chr14_-_12387102 | 2.12 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr3_-_162872831 | 1.57 |
ENSRNOT00000008478
|
Sulf2
|
sulfatase 2 |
| chr12_+_42097626 | 1.41 |
ENSRNOT00000001893
|
Tbx5
|
T-box 5 |
| chr5_+_63781801 | 1.39 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr3_+_55094637 | 1.37 |
ENSRNOT00000058763
|
Cers6
|
ceramide synthase 6 |
| chr1_-_82610350 | 1.25 |
ENSRNOT00000028177
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr1_+_201499028 | 1.23 |
ENSRNOT00000027860
|
Htra1
|
HtrA serine peptidase 1 |
| chr2_-_210550490 | 1.21 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr10_+_34692870 | 0.97 |
ENSRNOT00000048930
|
Olr1395
|
olfactory receptor 1395 |
| chr1_-_188895223 | 0.75 |
ENSRNOT00000032796
|
Gpr139
|
G protein-coupled receptor 139 |
| chr3_+_171832500 | 0.63 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr13_+_60619309 | 0.50 |
ENSRNOT00000082129
|
AABR07021204.1
|
|
| chr10_+_15191078 | 0.49 |
ENSRNOT00000026697
|
Wdr24
|
WD repeat domain 24 |
| chr5_-_155258392 | 0.30 |
ENSRNOT00000017065
|
C1qc
|
complement C1q C chain |
| chr10_+_74910713 | 0.08 |
ENSRNOT00000091619
|
Hsf5
|
heat shock transcription factor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zscan4c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 3.7 | 11.1 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 3.1 | 9.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 2.3 | 9.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 2.0 | 6.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.5 | 7.4 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 1.5 | 7.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 1.5 | 4.4 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 1.3 | 18.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.2 | 3.7 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.2 | 11.6 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 1.1 | 4.5 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 1.0 | 3.0 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 1.0 | 6.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.9 | 9.8 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.6 | 5.5 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.6 | 4.2 | GO:1900020 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.5 | 1.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.4 | 1.2 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.4 | 1.2 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 13.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 2.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 1.4 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 17.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 1.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 3.1 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 11.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.2 | 9.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.2 | 27.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 3.9 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 4.7 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 7.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 6.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 5.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.3 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 1.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 3.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.6 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 2.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 3.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.5 | 10.7 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 1.5 | 22.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.0 | 4.2 | GO:0044307 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.8 | 18.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.4 | 1.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 5.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 9.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 3.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 15.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 26.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 5.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 4.4 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 23.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 5.9 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 20.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 3.5 | 13.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 3.1 | 9.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 2.3 | 6.8 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.4 | 9.8 | GO:0043426 | MRF binding(GO:0043426) |
| 1.3 | 18.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.1 | 4.4 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.0 | 15.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.8 | 3.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.8 | 6.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.6 | 5.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.6 | 4.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 1.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.4 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 11.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 7.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 7.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.2 | 5.5 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.2 | 9.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 2.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 3.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 3.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 24.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 9.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 4.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 24.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 8.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 15.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 9.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 6.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 8.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 7.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 9.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 18.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 7.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 13.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.6 | 9.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.5 | 6.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 4.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 9.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 4.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 6.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 4.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |