Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zkscan3
Z-value: 0.90
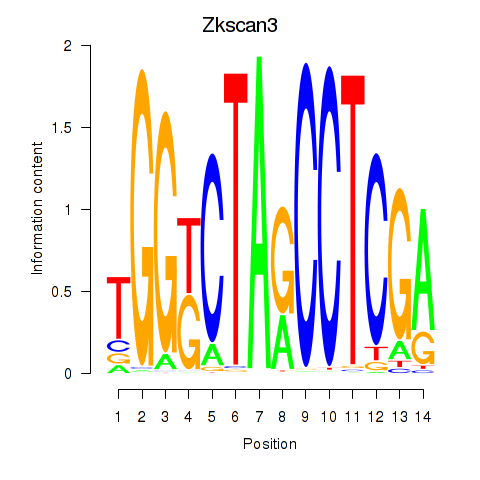
Transcription factors associated with Zkscan3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan3
|
ENSRNOG00000055000 | zinc finger with KRAB and SCAN domains 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan3 | rn6_v1_chr17_+_45247776_45247794 | -0.31 | 1.2e-08 | Click! |
Activity profile of Zkscan3 motif
Sorted Z-values of Zkscan3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_48365784 | 58.14 |
ENSRNOT00000077317
|
Dao
|
D-amino-acid oxidase |
| chr1_-_221917901 | 45.94 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr3_-_94686989 | 44.74 |
ENSRNOT00000016677
|
Depdc7
|
DEP domain containing 7 |
| chr10_-_62254287 | 33.44 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr9_-_82461903 | 29.99 |
ENSRNOT00000026654
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr9_+_49479023 | 29.08 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr4_-_148845267 | 28.41 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr15_-_45550285 | 27.06 |
ENSRNOT00000012948
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr9_-_71852113 | 25.28 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr12_+_47698947 | 22.23 |
ENSRNOT00000001586
|
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4 |
| chr10_+_14216155 | 22.20 |
ENSRNOT00000020192
|
Hagh
|
hydroxyacyl glutathione hydrolase |
| chr10_-_14215957 | 21.14 |
ENSRNOT00000019767
|
Fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr7_-_70452675 | 19.66 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr9_+_100281339 | 18.35 |
ENSRNOT00000029127
|
Agxt
|
alanine-glyoxylate aminotransferase |
| chr13_+_49074644 | 17.13 |
ENSRNOT00000000041
|
Klhdc8a
|
kelch domain containing 8A |
| chr19_-_43911057 | 15.46 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr7_+_11301957 | 13.91 |
ENSRNOT00000027847
|
Tjp3
|
tight junction protein 3 |
| chr4_-_161907767 | 13.71 |
ENSRNOT00000009557
|
A2ml1
|
alpha-2-macroglobulin-like 1 |
| chr11_-_57993548 | 13.65 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr12_-_39396042 | 13.30 |
ENSRNOT00000001746
|
P2rx7
|
purinergic receptor P2X 7 |
| chr2_+_197682000 | 11.77 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr7_+_64672722 | 11.38 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr7_-_140770647 | 10.95 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr4_+_157107469 | 9.75 |
ENSRNOT00000015678
|
C1rl
|
complement C1r subcomponent like |
| chr16_+_81665540 | 9.30 |
ENSRNOT00000026546
ENSRNOT00000092644 ENSRNOT00000073240 |
Grtp1
|
growth hormone regulated TBC protein 1 |
| chr20_+_41266566 | 8.61 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr12_-_2277169 | 8.48 |
ENSRNOT00000078078
|
Clec4g
|
C-type lectin domain family 4, member G |
| chr8_+_57886168 | 7.44 |
ENSRNOT00000039336
|
Exph5
|
exophilin 5 |
| chr1_+_99532568 | 6.79 |
ENSRNOT00000077073
|
Zfp819
|
zinc finger protein 819 |
| chr7_+_70452579 | 6.78 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr11_-_69355854 | 6.60 |
ENSRNOT00000002975
|
Ropn1
|
rhophilin associated tail protein 1 |
| chr1_+_78671121 | 6.22 |
ENSRNOT00000021310
|
Ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr13_-_72869396 | 6.04 |
ENSRNOT00000093563
|
RGD1304622
|
similar to 6820428L09 protein |
| chr8_+_71719563 | 5.59 |
ENSRNOT00000022828
|
Ppib
|
peptidylprolyl isomerase B |
| chr1_+_85252103 | 5.46 |
ENSRNOT00000080740
|
Ifnl1
|
interferon, lambda 1 |
| chr11_-_28478360 | 5.38 |
ENSRNOT00000032663
|
Cldn17
|
claudin 17 |
| chr19_-_44095250 | 4.77 |
ENSRNOT00000074824
|
Tmem170a
|
transmembrane protein 170A |
| chr19_-_52424318 | 4.70 |
ENSRNOT00000021743
|
Tldc1
|
TBC/LysM-associated domain containing 1 |
| chr19_-_39087880 | 4.61 |
ENSRNOT00000070822
|
Chtf8
|
chromosome transmission fidelity factor 8 |
| chr1_-_85237775 | 4.51 |
ENSRNOT00000035328
|
Ifnl3
|
interferon, lambda 3 |
| chr3_+_11679530 | 4.29 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr4_-_125808281 | 3.96 |
ENSRNOT00000037848
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_+_44506098 | 3.83 |
ENSRNOT00000078135
|
LOC501698
|
similar to olfactory receptor Olr1448 |
| chr9_-_99680979 | 3.80 |
ENSRNOT00000049347
|
Olr1345
|
olfactory receptor 1345 |
| chr4_-_7113919 | 3.61 |
ENSRNOT00000014082
|
Chpf2
|
chondroitin polymerizing factor 2 |
| chr10_+_20591432 | 3.18 |
ENSRNOT00000059780
|
Pank3
|
pantothenate kinase 3 |
| chr3_+_119783717 | 2.73 |
ENSRNOT00000039043
|
Astl
|
astacin like metalloendopeptidase |
| chrX_-_139229702 | 2.30 |
ENSRNOT00000042507
|
AABR07041771.1
|
|
| chr2_-_154508877 | 2.09 |
ENSRNOT00000086472
|
RGD1565059
|
similar to hypothetical protein E130311K13 |
| chr1_-_174620064 | 2.06 |
ENSRNOT00000016224
|
Tmem41b
|
transmembrane protein 41B |
| chr1_+_204861566 | 1.86 |
ENSRNOT00000023214
|
Fam175b
|
family with sequence similarity 175, member B |
| chr1_+_277190964 | 1.86 |
ENSRNOT00000080511
|
Casp7
|
caspase 7 |
| chr10_-_34961349 | 1.58 |
ENSRNOT00000004885
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr3_+_143084563 | 1.49 |
ENSRNOT00000006362
|
Cstl1
|
cystatin-like 1 |
| chr5_+_157434481 | 1.23 |
ENSRNOT00000088556
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr3_-_148104483 | 1.17 |
ENSRNOT00000055406
|
Defb25
|
defensin beta 25 |
| chr10_+_55169282 | 0.99 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr15_-_58711872 | 0.96 |
ENSRNOT00000058204
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr10_-_37055282 | 0.84 |
ENSRNOT00000086193
ENSRNOT00000065584 |
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr19_+_49462129 | 0.75 |
ENSRNOT00000015158
|
Cenpn
|
centromere protein N |
| chr14_-_89179507 | 0.69 |
ENSRNOT00000006498
|
Pkd1l1
|
polycystin 1 like 1, transient receptor potential channel interacting |
| chr10_+_105861743 | 0.54 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chrX_+_70256737 | 0.53 |
ENSRNOT00000029298
|
Otud6a
|
OTU deubiquitinase 6A |
| chr1_-_242373764 | 0.51 |
ENSRNOT00000076544
|
Fam122a
|
family with sequence similarity 122A |
| chr6_-_117972898 | 0.02 |
ENSRNOT00000032968
|
AABR07065265.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Zkscan3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.4 | 58.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 7.4 | 22.2 | GO:0072566 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 7.3 | 29.1 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 6.1 | 18.4 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) oxalic acid secretion(GO:0046724) |
| 4.4 | 22.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 4.4 | 13.3 | GO:0043132 | phospholipid transfer to membrane(GO:0006649) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) |
| 4.2 | 45.9 | GO:0015747 | urate transport(GO:0015747) |
| 3.7 | 33.4 | GO:0071279 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 2.0 | 30.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 1.9 | 5.6 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 1.7 | 11.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.5 | 13.7 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 1.4 | 4.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 1.4 | 5.5 | GO:0042509 | negative regulation of interleukin-13 production(GO:0032696) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 1.4 | 6.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.9 | 2.7 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.5 | 4.6 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 | 27.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.5 | 8.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.4 | 7.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 1.9 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.3 | 4.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 9.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 6.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 11.0 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.2 | 15.5 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 0.8 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.2 | 1.9 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 4.7 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.2 | 4.5 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.2 | 0.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 3.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 9.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 8.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 15.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.7 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 13.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.5 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 4.0 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 1.6 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 33.4 | GO:0043203 | axon hillock(GO:0043203) |
| 1.6 | 6.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 1.5 | 13.3 | GO:0032059 | bleb(GO:0032059) |
| 1.1 | 6.6 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 1.1 | 27.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.9 | 18.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.9 | 58.1 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.5 | 4.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 5.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 45.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 20.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 17.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 11.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 36.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.2 | 30.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 6.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 11.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.6 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.1 | 22.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 8.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 56.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.0 | 21.1 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 4.6 | 45.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 4.4 | 22.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 3.7 | 18.4 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 2.5 | 58.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 2.5 | 22.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.5 | 13.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.1 | 29.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.1 | 27.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.9 | 4.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.8 | 3.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.6 | 30.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 11.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.5 | 4.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 10.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.4 | 8.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 33.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 15.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.5 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 8.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 5.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 25.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 11.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 17.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 5.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 22.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 38.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 4.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 19.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 45.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.1 | 13.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 15.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 6.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 76.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 44.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 3.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 5.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 5.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |