Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zkscan1
Z-value: 0.65
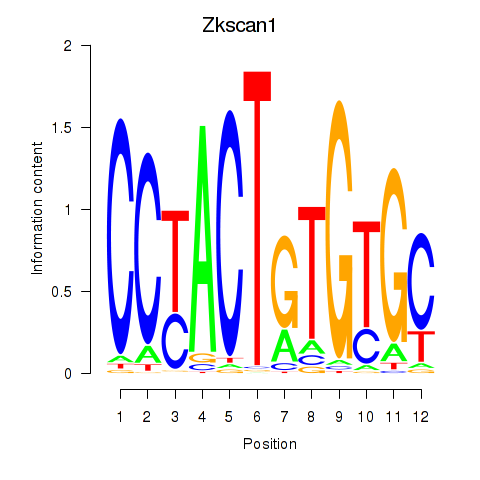
Transcription factors associated with Zkscan1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan1
|
ENSRNOG00000001335 | zinc finger with KRAB and SCAN domains 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan1 | rn6_v1_chr12_+_19231092_19231092 | 0.43 | 5.3e-16 | Click! |
Activity profile of Zkscan1 motif
Sorted Z-values of Zkscan1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_143059764 | 20.88 |
ENSRNOT00000010660
ENSRNOT00000087370 |
Krt7
|
keratin 7 |
| chr14_+_1462358 | 20.60 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr8_+_55603968 | 18.77 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr10_-_65692016 | 17.72 |
ENSRNOT00000085074
ENSRNOT00000038690 |
Slc13a2
|
solute carrier family 13 member 2 |
| chr7_-_119797098 | 16.36 |
ENSRNOT00000009994
|
Rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr1_+_89220083 | 15.77 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr5_-_137112927 | 12.92 |
ENSRNOT00000078302
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr5_+_151413382 | 12.77 |
ENSRNOT00000012626
|
Cd164l2
|
CD164 molecule like 2 |
| chr15_+_31642169 | 12.23 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr2_-_219097619 | 12.16 |
ENSRNOT00000078806
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr5_+_154522119 | 12.07 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr7_-_123767797 | 11.94 |
ENSRNOT00000012699
|
Tcf20
|
transcription factor 20 |
| chr10_-_65424802 | 11.65 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr12_+_943006 | 11.39 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chr3_-_1584946 | 11.35 |
ENSRNOT00000031058
|
Pax8
|
paired box 8 |
| chr6_-_142585188 | 10.97 |
ENSRNOT00000067437
|
AABR07065815.1
|
|
| chr6_-_138632159 | 10.88 |
ENSRNOT00000082921
ENSRNOT00000040702 |
Ighm
|
immunoglobulin heavy constant mu |
| chr13_+_90244681 | 10.37 |
ENSRNOT00000078162
|
Cd84
|
CD84 molecule |
| chr6_-_142635763 | 10.31 |
ENSRNOT00000048908
|
AABR07065815.2
|
|
| chr12_-_36587839 | 10.16 |
ENSRNOT00000001295
|
Bri3bp
|
Bri3 binding protein |
| chr15_-_29446332 | 9.88 |
ENSRNOT00000082901
|
AABR07017634.1
|
|
| chr7_-_126913585 | 9.53 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr3_-_151625644 | 8.72 |
ENSRNOT00000026657
|
Rbm12
|
RNA binding motif protein 12 |
| chr6_+_139486775 | 8.68 |
ENSRNOT00000077771
|
AABR07065699.3
|
|
| chr18_-_69944632 | 8.64 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr8_+_67295727 | 8.59 |
ENSRNOT00000020443
|
Anp32a
|
acidic nuclear phosphoprotein 32 family member A |
| chr16_-_39476384 | 8.47 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr7_-_116504853 | 8.38 |
ENSRNOT00000056557
|
RGD1565410
|
similar to Ly6-C antigen gene |
| chr5_-_159602251 | 8.31 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr1_-_262014066 | 8.11 |
ENSRNOT00000087083
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr19_-_55257876 | 7.85 |
ENSRNOT00000017564
|
Cyba
|
cytochrome b-245 alpha chain |
| chr16_-_39476025 | 7.34 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr6_-_140102325 | 7.13 |
ENSRNOT00000072238
|
AABR07065750.2
|
|
| chr14_-_45165207 | 6.72 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr8_-_120446455 | 6.63 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr5_+_34040258 | 6.48 |
ENSRNOT00000009758
|
Ggh
|
gamma-glutamyl hydrolase |
| chr14_-_45859908 | 6.44 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr7_+_18409147 | 6.36 |
ENSRNOT00000086336
ENSRNOT00000011849 |
Adamts10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr14_-_5378726 | 6.27 |
ENSRNOT00000002896
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr7_-_141001993 | 6.04 |
ENSRNOT00000092068
|
Fmnl3
|
formin-like 3 |
| chr3_-_148312420 | 5.86 |
ENSRNOT00000047416
ENSRNOT00000081272 |
Bcl2l1
|
Bcl2-like 1 |
| chr4_+_57378069 | 5.73 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr6_-_102196138 | 5.65 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr2_-_30577218 | 5.60 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr1_-_87147308 | 5.48 |
ENSRNOT00000027773
ENSRNOT00000089305 ENSRNOT00000090402 |
Actn4
|
actinin alpha 4 |
| chrX_-_37705263 | 5.25 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr7_-_118840634 | 5.04 |
ENSRNOT00000031568
|
Apol11a
|
apolipoprotein L 11a |
| chr5_-_153252021 | 5.01 |
ENSRNOT00000023221
|
Tmem50a
|
transmembrane protein 50A |
| chr17_+_9282675 | 4.99 |
ENSRNOT00000051702
|
H2afy
|
H2A histone family, member Y |
| chr3_-_80875817 | 4.74 |
ENSRNOT00000091265
|
Dgkz
|
diacylglycerol kinase zeta |
| chr1_+_220869805 | 4.70 |
ENSRNOT00000015962
|
Cfl1
|
cofilin 1 |
| chr5_-_2803855 | 4.56 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr15_+_18399733 | 4.41 |
ENSRNOT00000061158
|
Fam107a
|
family with sequence similarity 107, member A |
| chr7_+_11301957 | 4.32 |
ENSRNOT00000027847
|
Tjp3
|
tight junction protein 3 |
| chr1_-_204817080 | 4.28 |
ENSRNOT00000077956
|
Fam53b
|
family with sequence similarity 53, member B |
| chr3_-_148312791 | 4.19 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr7_-_23594133 | 4.15 |
ENSRNOT00000005746
|
Timp3
|
TIMP metallopeptidase inhibitor 3 |
| chr7_-_12673659 | 4.12 |
ENSRNOT00000091650
ENSRNOT00000041277 ENSRNOT00000044865 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr16_+_71738718 | 4.09 |
ENSRNOT00000022097
|
Plekha2
|
pleckstrin homology domain containing A2 |
| chr6_-_138631997 | 3.93 |
ENSRNOT00000073304
|
AABR07065651.3
|
|
| chr10_+_71278650 | 3.88 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr4_+_77554269 | 3.80 |
ENSRNOT00000037248
|
Zfp282
|
zinc finger protein 282 |
| chr1_+_219000844 | 3.71 |
ENSRNOT00000022486
|
Kmt5b
|
lysine methyltransferase 5B |
| chr8_-_111965889 | 3.49 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr3_+_152909189 | 3.47 |
ENSRNOT00000066341
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr17_-_88140588 | 3.37 |
ENSRNOT00000006662
|
RGD1566369
|
similar to ribosomal protein S8 |
| chr3_+_72134731 | 3.22 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr5_-_151473750 | 3.18 |
ENSRNOT00000011357
|
Tmem222
|
transmembrane protein 222 |
| chr13_-_82753438 | 3.16 |
ENSRNOT00000075948
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr1_-_262013619 | 3.10 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr3_-_10602672 | 3.04 |
ENSRNOT00000011648
|
Ncs1
|
neuronal calcium sensor 1 |
| chr12_+_25093149 | 3.02 |
ENSRNOT00000050059
|
Eif4h
|
eukaryotic translation initiation factor 4H |
| chr1_-_228755866 | 2.99 |
ENSRNOT00000083283
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chrX_+_31140960 | 2.96 |
ENSRNOT00000084395
ENSRNOT00000004494 |
Mospd2
|
motile sperm domain containing 2 |
| chr6_+_126623015 | 2.92 |
ENSRNOT00000010672
|
Tmem251
|
transmembrane protein 251 |
| chr15_-_18695133 | 2.88 |
ENSRNOT00000012271
|
Abhd6
|
abhydrolase domain containing 6 |
| chr4_+_87293871 | 2.86 |
ENSRNOT00000090943
|
AABR07060628.1
|
|
| chr8_+_116804451 | 2.70 |
ENSRNOT00000041402
|
Ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr1_+_203971152 | 2.61 |
ENSRNOT00000075540
|
Gpr26
|
G protein-coupled receptor 26 |
| chr7_-_75288365 | 2.61 |
ENSRNOT00000036904
|
Ankrd46
|
ankyrin repeat domain 46 |
| chr10_+_71217966 | 2.60 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr17_+_20619324 | 2.54 |
ENSRNOT00000079788
|
AABR07027235.1
|
|
| chr8_-_28075514 | 2.49 |
ENSRNOT00000072851
|
Vps26b
|
VPS26 retromer complex component B |
| chr8_-_115358046 | 2.37 |
ENSRNOT00000017607
|
Grm2
|
glutamate metabotropic receptor 2 |
| chr3_+_119805941 | 2.36 |
ENSRNOT00000018584
|
Adra2b
|
adrenoceptor alpha 2B |
| chr1_-_101054145 | 2.33 |
ENSRNOT00000082560
|
Prrg2
|
proline rich and Gla domain 2 |
| chr6_+_10674371 | 2.24 |
ENSRNOT00000020337
|
Socs5
|
suppressor of cytokine signaling 5 |
| chr7_-_73130740 | 2.10 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr8_+_19888667 | 2.09 |
ENSRNOT00000078593
|
Zfp317
|
zinc finger protein 317 |
| chr8_-_21995806 | 1.98 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr18_+_29591611 | 1.91 |
ENSRNOT00000043271
|
Wdr55
|
WD repeat domain 55 |
| chr13_-_97282299 | 1.82 |
ENSRNOT00000004244
|
Mixl1
|
Mix paired-like homeobox 1 |
| chr17_-_10818835 | 1.77 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr19_-_38484611 | 1.76 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr16_+_64729221 | 1.73 |
ENSRNOT00000015297
|
Mak16
|
MAK16 homolog |
| chr14_+_113202419 | 1.58 |
ENSRNOT00000004764
|
Efemp1
|
EGF-containing fibulin-like extracellular matrix protein 1 |
| chr5_+_148320438 | 1.35 |
ENSRNOT00000018742
|
Pef1
|
penta-EF hand domain containing 1 |
| chr17_-_57816256 | 1.28 |
ENSRNOT00000051570
|
LOC103694120
|
isopentenyl-diphosphate delta-isomerase 2-like |
| chr5_+_171297850 | 1.21 |
ENSRNOT00000034284
|
Lrrc47
|
leucine rich repeat containing 47 |
| chr3_-_10226286 | 1.20 |
ENSRNOT00000093627
|
Fubp3
|
far upstream element binding protein 3 |
| chr1_+_274030978 | 0.94 |
ENSRNOT00000076387
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr4_+_57034675 | 0.92 |
ENSRNOT00000080223
|
Smo
|
smoothened, frizzled class receptor |
| chr10_+_110469290 | 0.89 |
ENSRNOT00000054919
|
Foxk2
|
forkhead box K2 |
| chr4_-_85915099 | 0.88 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr10_-_104718621 | 0.83 |
ENSRNOT00000011560
|
Fbf1
|
Fas binding factor 1 |
| chr4_-_27755103 | 0.83 |
ENSRNOT00000012253
|
Fam133b
|
family with sequence similarity 133, member B |
| chr6_-_29999410 | 0.74 |
ENSRNOT00000075790
|
Sf3b6
|
splicing factor 3B, subunit 6 |
| chr17_+_70684340 | 0.69 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr12_-_2540668 | 0.68 |
ENSRNOT00000001398
|
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr17_+_26785029 | 0.63 |
ENSRNOT00000022065
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr17_+_70684539 | 0.63 |
ENSRNOT00000025700
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr6_+_69971227 | 0.60 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr9_-_49950093 | 0.56 |
ENSRNOT00000023014
|
Fhl2
|
four and a half LIM domains 2 |
| chr15_-_12513931 | 0.56 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr8_+_128044084 | 0.55 |
ENSRNOT00000019106
|
Xylb
|
xylulokinase |
| chr6_+_93281408 | 0.54 |
ENSRNOT00000009610
|
Frmd6
|
FERM domain containing 6 |
| chr5_-_169331163 | 0.24 |
ENSRNOT00000042301
|
Espn
|
espin |
| chr7_+_70292565 | 0.21 |
ENSRNOT00000073237
|
Avil
|
advillin |
| chr1_-_61686944 | 0.13 |
ENSRNOT00000059638
|
Vom1r23
|
vomeronasal 1 receptor 23 |
| chr6_+_111642411 | 0.12 |
ENSRNOT00000016962
|
Adck1
|
aarF domain containing kinase 1 |
| chr15_-_28733513 | 0.04 |
ENSRNOT00000078180
|
Sall2
|
spalt-like transcription factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zkscan1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 20.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 3.9 | 15.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 3.8 | 11.3 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 3.4 | 10.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 3.2 | 9.5 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 2.6 | 7.9 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 2.6 | 10.4 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 2.2 | 12.9 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 2.0 | 16.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 2.0 | 12.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.7 | 5.0 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 1.6 | 4.7 | GO:2000769 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 1.4 | 11.4 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 1.4 | 11.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 1.2 | 12.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 1.1 | 5.5 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 1.1 | 11.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 1.1 | 3.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 1.0 | 4.1 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.0 | 3.0 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.9 | 2.6 | GO:0061235 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.7 | 2.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.6 | 2.4 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.6 | 2.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.6 | 5.6 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.5 | 3.7 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.5 | 4.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 1.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.4 | 8.6 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.4 | 16.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 2.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 4.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 3.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 1.8 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 5.7 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.2 | 0.9 | GO:0061113 | positive regulation of hh target transcription factor activity(GO:0007228) pancreas morphogenesis(GO:0061113) |
| 0.2 | 3.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 2.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 0.5 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.2 | 1.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 1.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 4.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 6.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 24.0 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 1.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 4.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.6 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 2.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.9 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 3.8 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 6.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 4.3 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 1.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 2.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 3.0 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.7 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 6.7 | GO:1901653 | cellular response to peptide(GO:1901653) |
| 0.0 | 24.8 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 1.6 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 5.0 | GO:0006897 | endocytosis(GO:0006897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 2.2 | 11.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 1.4 | 10.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.8 | 7.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 5.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.6 | 6.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 1.8 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.4 | 12.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.4 | 3.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 5.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 16.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 3.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 4.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 5.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 18.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.7 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 16.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 8.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 12.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 6.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 4.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 14.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 4.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 36.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 4.1 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 11.7 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 26.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 25.0 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 3.8 | 11.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 3.2 | 12.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.7 | 10.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.4 | 4.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.3 | 6.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.2 | 3.7 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.1 | 12.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.0 | 3.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.9 | 2.7 | GO:0052724 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.9 | 7.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 5.0 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.8 | 2.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.8 | 2.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.8 | 3.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.6 | 6.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.6 | 11.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 6.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 1.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 4.7 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.4 | 18.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 3.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.3 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.3 | 8.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 6.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 2.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 3.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 2.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 4.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 3.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 5.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 24.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 17.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 8.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 13.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 9.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 6.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.7 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 11.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 16.1 | GO:0001159 | core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 14.5 | GO:0005198 | structural molecule activity(GO:0005198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 32.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.8 | 16.8 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.6 | 10.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 16.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 11.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 7.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 5.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 8.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 5.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 5.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 17.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 12.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.5 | 20.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.5 | 10.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.5 | 16.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.4 | 11.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.4 | 5.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 4.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 6.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 5.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 12.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 7.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 3.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 8.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 6.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 3.0 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 2.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 4.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |