Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zic4
Z-value: 1.01
Transcription factors associated with Zic4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
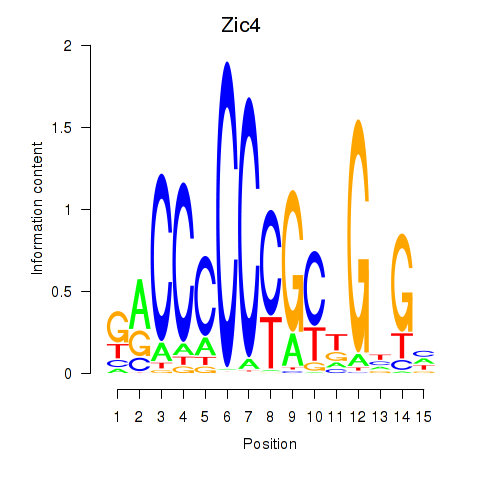
Zic4
|
ENSRNOG00000014871 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic4 | rn6_v1_chr8_+_98755104_98755104 | 0.63 | 2.2e-37 | Click! |
Activity profile of Zic4 motif
Sorted Z-values of Zic4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24923128 | 69.15 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr3_-_148428494 | 38.43 |
ENSRNOT00000011350
|
Dusp15
|
dual specificity phosphatase 15 |
| chr3_-_148428288 | 32.70 |
ENSRNOT00000072663
|
Dusp15
|
dual specificity phosphatase 15 |
| chr5_+_156876706 | 31.08 |
ENSRNOT00000021864
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr9_+_82120059 | 31.06 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr10_-_91047177 | 31.02 |
ENSRNOT00000003986
|
C1ql1
|
complement C1q like 1 |
| chr1_+_266953139 | 29.18 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr5_-_40237591 | 27.57 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr19_-_9801942 | 25.87 |
ENSRNOT00000051414
ENSRNOT00000017494 |
Ndrg4
|
NDRG family member 4 |
| chr8_-_36760742 | 23.67 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr17_-_10208360 | 22.95 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr10_+_80790168 | 21.98 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr7_+_140788987 | 21.79 |
ENSRNOT00000086611
|
Kcnh3
|
potassium voltage-gated channel subfamily H member 3 |
| chr1_+_84470829 | 21.74 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr1_-_212549477 | 21.32 |
ENSRNOT00000024765
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr5_+_123905166 | 20.93 |
ENSRNOT00000082021
|
Dab1
|
DAB1, reelin adaptor protein |
| chr12_+_36694960 | 20.81 |
ENSRNOT00000064276
ENSRNOT00000001299 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr17_+_9109731 | 19.32 |
ENSRNOT00000016009
|
Cxcl14
|
C-X-C motif chemokine ligand 14 |
| chrX_-_157312028 | 18.98 |
ENSRNOT00000077979
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chrX_-_82986051 | 18.57 |
ENSRNOT00000077587
|
Hdx
|
highly divergent homeobox |
| chr2_+_149214265 | 18.01 |
ENSRNOT00000084020
|
Med12l
|
mediator complex subunit 12-like |
| chr7_-_107009330 | 17.59 |
ENSRNOT00000074573
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr10_-_92008082 | 17.16 |
ENSRNOT00000006361
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr1_-_112811936 | 17.15 |
ENSRNOT00000093339
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr19_+_20607507 | 16.78 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr13_+_110920830 | 16.49 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr3_+_3310954 | 16.47 |
ENSRNOT00000061773
|
Kcnt1
|
potassium sodium-activated channel subfamily T member 1 |
| chr1_-_216663720 | 16.33 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr8_-_103190243 | 15.62 |
ENSRNOT00000075305
|
Chst2
|
carbohydrate sulfotransferase 2 |
| chr2_-_211207465 | 15.44 |
ENSRNOT00000027263
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chrX_-_136807885 | 15.35 |
ENSRNOT00000010325
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr1_+_266952561 | 15.16 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr19_-_15431274 | 14.66 |
ENSRNOT00000022285
ENSRNOT00000048470 |
Slc6a2
|
solute carrier family 6 member 2 |
| chr13_-_51076165 | 14.50 |
ENSRNOT00000004602
|
Adora1
|
adenosine A1 receptor |
| chr19_-_43596801 | 13.93 |
ENSRNOT00000025625
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr10_-_40953651 | 13.79 |
ENSRNOT00000063889
|
Glra1
|
glycine receptor, alpha 1 |
| chr1_-_199270627 | 13.75 |
ENSRNOT00000026063
|
Stx1b
|
syntaxin 1B |
| chr3_+_112916217 | 13.25 |
ENSRNOT00000030065
|
Tmem62
|
transmembrane protein 62 |
| chr3_-_2803574 | 12.72 |
ENSRNOT00000040995
|
RGD1560470
|
similar to Gene model 996 |
| chr16_+_26739389 | 12.52 |
ENSRNOT00000047223
|
Klhl2
|
kelch-like family member 2 |
| chr5_-_88629491 | 12.37 |
ENSRNOT00000058906
|
Tle1
|
transducin like enhancer of split 1 |
| chr16_+_74886719 | 11.32 |
ENSRNOT00000089265
|
Atp7b
|
ATPase copper transporting beta |
| chr5_+_135962911 | 10.78 |
ENSRNOT00000087353
|
Ptch2
|
patched 2 |
| chr1_-_84491466 | 10.66 |
ENSRNOT00000034609
|
Map3k10
|
mitogen activated protein kinase kinase kinase 10 |
| chr20_-_1984737 | 10.59 |
ENSRNOT00000040232
ENSRNOT00000051634 ENSRNOT00000079445 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr10_-_40953467 | 10.22 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr20_-_3822754 | 10.07 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr4_+_132137793 | 10.04 |
ENSRNOT00000014455
|
Gpr27
|
G protein-coupled receptor 27 |
| chr1_-_78997869 | 9.89 |
ENSRNOT00000023490
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr11_+_75434197 | 9.70 |
ENSRNOT00000032569
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr17_-_31535172 | 9.51 |
ENSRNOT00000023914
|
Bphl
|
biphenyl hydrolase like |
| chr13_+_71107465 | 9.14 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr16_+_10267482 | 8.92 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr7_-_124620703 | 8.80 |
ENSRNOT00000017727
|
Scube1
|
signal peptide, CUB domain and EGF like domain containing 1 |
| chr2_-_26438790 | 8.79 |
ENSRNOT00000035017
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr1_+_226435979 | 8.52 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr5_-_150323063 | 8.36 |
ENSRNOT00000014084
|
Oprd1
|
opioid receptor, delta 1 |
| chr10_+_48240127 | 7.75 |
ENSRNOT00000080682
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_+_219348721 | 7.44 |
ENSRNOT00000025084
|
Pitpnm1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr6_-_6842758 | 7.21 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr20_-_31598118 | 7.06 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr8_+_117246376 | 6.98 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr17_-_9695292 | 6.94 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr10_-_109840047 | 6.75 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr12_+_25036605 | 6.19 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr2_+_95008311 | 6.09 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr5_-_142845116 | 5.96 |
ENSRNOT00000065105
|
RGD1559909
|
RGD1559909 |
| chr2_+_95008477 | 5.88 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr7_-_139254149 | 5.70 |
ENSRNOT00000078644
|
Rapgef3
|
Rap guanine nucleotide exchange factor 3 |
| chr6_+_33885495 | 5.62 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr16_-_20686317 | 5.48 |
ENSRNOT00000060097
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr16_+_59988603 | 5.26 |
ENSRNOT00000015176
|
Cldn23
|
claudin 23 |
| chr8_+_45797315 | 5.23 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr5_-_28131133 | 5.21 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr1_-_221605400 | 5.03 |
ENSRNOT00000028542
|
Ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr1_-_165967069 | 4.86 |
ENSRNOT00000089359
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr10_+_48240330 | 4.85 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr3_+_175885894 | 4.57 |
ENSRNOT00000039780
|
Ntsr1
|
neurotensin receptor 1 |
| chr17_-_52569036 | 4.46 |
ENSRNOT00000019396
|
Gli3
|
GLI family zinc finger 3 |
| chr5_-_74190991 | 4.46 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr7_-_117786872 | 4.44 |
ENSRNOT00000030517
|
Lrrc24
|
leucine rich repeat containing 24 |
| chr5_-_144479306 | 4.02 |
ENSRNOT00000087697
|
Ago1
|
argonaute 1, RISC catalytic component |
| chr10_-_75270561 | 3.88 |
ENSRNOT00000012279
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr6_+_69950500 | 3.76 |
ENSRNOT00000049500
|
RGD1559629
|
similar to H+ ATP synthase |
| chr16_+_59197367 | 3.74 |
ENSRNOT00000014924
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr6_-_126582034 | 3.69 |
ENSRNOT00000010656
ENSRNOT00000080829 |
Itpk1
|
inositol-tetrakisphosphate 1-kinase |
| chr3_+_147226004 | 3.57 |
ENSRNOT00000012765
|
Tmem74b
|
transmembrane protein 74B |
| chr19_+_39229754 | 3.39 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr5_-_28130803 | 3.38 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr5_-_173622645 | 3.27 |
ENSRNOT00000045678
|
Agrn
|
agrin |
| chr20_-_422464 | 3.27 |
ENSRNOT00000051646
|
Olr1673
|
olfactory receptor 1673 |
| chr10_+_86950557 | 3.26 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr18_+_3995921 | 3.17 |
ENSRNOT00000091424
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr4_-_113886994 | 3.05 |
ENSRNOT00000037333
|
Htra2
|
HtrA serine peptidase 2 |
| chr6_+_104017607 | 2.84 |
ENSRNOT00000033378
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr7_-_144960527 | 2.84 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr10_+_15485905 | 2.70 |
ENSRNOT00000027609
|
Tmem8a
|
transmembrane protein 8A |
| chr5_-_74191167 | 2.58 |
ENSRNOT00000088169
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr6_+_1657331 | 2.52 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr14_-_84170301 | 2.52 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr10_-_13898855 | 2.42 |
ENSRNOT00000004249
|
Rab26
|
RAB26, member RAS oncogene family |
| chr3_+_151609602 | 2.41 |
ENSRNOT00000065052
|
Spag4
|
sperm associated antigen 4 |
| chr5_+_147714163 | 2.33 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr1_-_67390141 | 2.26 |
ENSRNOT00000025808
|
Sbk1
|
SH3 domain binding kinase 1 |
| chr1_+_146037426 | 2.21 |
ENSRNOT00000055583
|
Mesdc2
|
mesoderm development candidate 2 |
| chr7_+_12006710 | 2.16 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr7_+_54031316 | 2.16 |
ENSRNOT00000039096
|
Bbs10
|
Bardet-Biedl syndrome 10 |
| chr1_-_277827178 | 2.14 |
ENSRNOT00000084915
ENSRNOT00000073346 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr3_+_147585947 | 2.05 |
ENSRNOT00000006833
|
Scrt2
|
scratch family transcriptional repressor 2 |
| chrX_-_14331486 | 2.03 |
ENSRNOT00000067603
|
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr1_+_197659187 | 1.68 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr13_-_105141030 | 1.64 |
ENSRNOT00000003313
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr4_+_49369296 | 1.62 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr3_-_163814799 | 1.44 |
ENSRNOT00000055127
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr14_-_12387102 | 1.38 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr1_+_41325462 | 1.35 |
ENSRNOT00000081017
ENSRNOT00000078494 ENSRNOT00000088168 |
Esr1
|
estrogen receptor 1 |
| chr4_-_77747070 | 1.34 |
ENSRNOT00000009247
|
Zfp746
|
zinc finger protein 746 |
| chr15_+_24254042 | 0.94 |
ENSRNOT00000092161
|
Fbxo34
|
F-box protein 34 |
| chr2_-_154508641 | 0.85 |
ENSRNOT00000065346
|
RGD1565059
|
similar to hypothetical protein E130311K13 |
| chr7_+_117963740 | 0.74 |
ENSRNOT00000075405
|
LOC108348189
|
COMM domain-containing protein 5 |
| chr5_-_169167831 | 0.73 |
ENSRNOT00000012407
|
Phf13
|
PHD finger protein 13 |
| chr3_+_73010786 | 0.26 |
ENSRNOT00000051010
|
Olr447
|
olfactory receptor 447 |
| chr13_-_48284990 | 0.23 |
ENSRNOT00000086928
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr1_+_204861566 | 0.18 |
ENSRNOT00000023214
|
Fam175b
|
family with sequence similarity 175, member B |
| chr14_+_81362618 | 0.15 |
ENSRNOT00000017386
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr20_-_7397448 | 0.15 |
ENSRNOT00000059426
|
LOC294154
|
similar to chromosome 6 open reading frame 106 isoform a |
| chr7_+_116943057 | 0.09 |
ENSRNOT00000056527
|
Tigd5
|
tigger transposable element derived 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.8 | 69.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 7.8 | 31.1 | GO:0021586 | pons maturation(GO:0021586) |
| 7.0 | 20.9 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 6.9 | 20.8 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 6.2 | 31.0 | GO:0061743 | motor learning(GO:0061743) |
| 5.5 | 38.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 4.8 | 14.5 | GO:0042323 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 4.6 | 13.8 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 4.3 | 17.2 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 3.8 | 23.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 3.8 | 11.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 3.3 | 10.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 3.2 | 19.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 3.2 | 44.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 3.1 | 15.4 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 3.0 | 24.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 2.9 | 25.9 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 2.8 | 8.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 2.7 | 16.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 2.4 | 19.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 2.0 | 27.6 | GO:0036065 | fucosylation(GO:0036065) |
| 1.8 | 10.6 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 1.6 | 9.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 1.5 | 4.6 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 1.4 | 5.6 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.2 | 7.3 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 1.2 | 16.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.1 | 3.4 | GO:0090611 | multivesicular body assembly(GO:0036258) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.1 | 4.5 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 1.1 | 5.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.1 | 3.3 | GO:0045887 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 1.0 | 4.0 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 1.0 | 17.6 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 1.0 | 14.7 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) |
| 0.9 | 8.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.8 | 2.5 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.8 | 9.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.8 | 15.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.7 | 8.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.7 | 5.0 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.7 | 23.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.7 | 2.8 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.7 | 12.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.6 | 7.0 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.6 | 71.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.6 | 23.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.6 | 6.7 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.5 | 7.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 8.6 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.5 | 3.1 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.5 | 8.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.4 | 17.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 10.1 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.3 | 1.4 | GO:1990375 | baculum development(GO:1990375) |
| 0.3 | 2.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 3.7 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 6.2 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.2 | 3.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 1.6 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 | 3.7 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 31.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 2.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 4.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 2.1 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 | 7.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 2.8 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 12.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 3.9 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.2 | GO:0021815 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 4.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 9.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 2.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 2.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.0 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 2.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 1.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 5.5 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 31.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 7.8 | 31.0 | GO:0044301 | climbing fiber(GO:0044301) |
| 3.5 | 10.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.9 | 5.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 1.8 | 5.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.4 | 13.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.2 | 40.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 1.2 | 23.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 31.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.9 | 32.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.9 | 8.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.8 | 20.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.8 | 41.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.8 | 4.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.7 | 17.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.6 | 4.0 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.5 | 35.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.5 | 22.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.5 | 3.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.4 | 3.9 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.4 | 11.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 8.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 3.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 17.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.3 | 3.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 8.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 29.6 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.2 | 5.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 12.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 6.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 28.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 19.6 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 17.3 | GO:0098791 | trans-Golgi network(GO:0005802) Golgi subcompartment(GO:0098791) |
| 0.1 | 12.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 8.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 14.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 54.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 2.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 24.0 | GO:0030977 | taurine binding(GO:0030977) |
| 7.8 | 31.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 6.9 | 20.8 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 5.5 | 38.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 4.6 | 27.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 4.6 | 23.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 3.7 | 14.7 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 3.6 | 21.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 3.0 | 54.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 2.9 | 14.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 2.7 | 10.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 2.6 | 10.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.6 | 15.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 2.3 | 11.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 2.2 | 15.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.4 | 8.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.3 | 71.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 1.2 | 7.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.2 | 16.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.1 | 4.6 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.0 | 17.2 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 1.0 | 31.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.9 | 5.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.9 | 19.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.8 | 37.7 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.7 | 17.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.6 | 5.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.6 | 2.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.6 | 8.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 8.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 16.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.6 | 2.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.5 | 19.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.5 | 3.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 22.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.4 | 1.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 3.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 3.7 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.3 | 10.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 23.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.3 | 13.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 4.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 13.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 7.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 10.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 3.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 3.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 8.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 4.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 9.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 3.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 9.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 13.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 2.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 5.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 6.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.2 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 7.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 6.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 8.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 15.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 4.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 52.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.6 | 12.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.6 | 23.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.5 | 9.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.4 | 10.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 5.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 5.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 8.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 3.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 7.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 53.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 8.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 20.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 6.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 23.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.5 | 41.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 1.2 | 20.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.9 | 48.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.9 | 17.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.9 | 14.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.8 | 19.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.7 | 13.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.6 | 14.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 10.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 15.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.5 | 5.6 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.5 | 9.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.5 | 10.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 5.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.4 | 53.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.3 | 12.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.3 | 5.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 11.3 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.3 | 6.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 3.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 3.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 3.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 4.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 4.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 9.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 5.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |