Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zic3
Z-value: 0.79
Transcription factors associated with Zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic3
|
ENSRNOG00000000861 | Zic family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | rn6_v1_chrX_+_140878216_140878216 | -0.33 | 8.1e-10 | Click! |
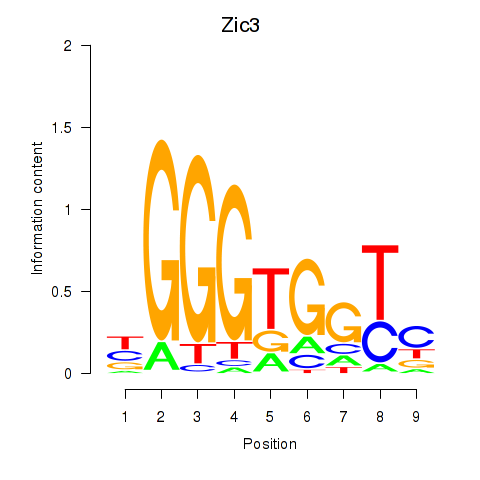
Activity profile of Zic3 motif
Sorted Z-values of Zic3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_7007287 | 41.81 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_-_7007051 | 36.47 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr7_+_28066635 | 30.15 |
ENSRNOT00000005844
|
Pah
|
phenylalanine hydroxylase |
| chr3_-_161212188 | 22.70 |
ENSRNOT00000065751
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr16_+_10267482 | 18.89 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr1_+_226687258 | 18.51 |
ENSRNOT00000079679
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr11_+_74057361 | 14.74 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr15_-_45524582 | 13.16 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr1_-_146736261 | 13.02 |
ENSRNOT00000068167
|
Fah
|
fumarylacetoacetate hydrolase |
| chr1_+_282568287 | 12.85 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr17_+_43458553 | 12.28 |
ENSRNOT00000088939
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr1_+_72636959 | 11.92 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr16_-_48981980 | 11.58 |
ENSRNOT00000014235
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr17_+_43460782 | 11.41 |
ENSRNOT00000059494
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr10_-_13580821 | 11.36 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chrX_+_111122552 | 10.38 |
ENSRNOT00000083566
ENSRNOT00000085078 ENSRNOT00000090928 |
Cldn2
|
claudin 2 |
| chr3_+_110367939 | 9.90 |
ENSRNOT00000010406
|
Bub1b
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr7_-_3342491 | 8.95 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr2_-_260596777 | 8.73 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr13_+_52624878 | 8.69 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr7_-_11777503 | 8.46 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr1_+_151918571 | 8.17 |
ENSRNOT00000022342
|
Ctsc
|
cathepsin C |
| chr1_+_261291870 | 7.73 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr6_+_27975417 | 7.73 |
ENSRNOT00000077830
|
Dtnb
|
dystrobrevin, beta |
| chr4_-_82289672 | 7.22 |
ENSRNOT00000036394
|
Hoxa10
|
homeobox A10 |
| chr1_-_90956544 | 6.95 |
ENSRNOT00000051422
|
LOC108348128
|
nesprin-4 |
| chr1_+_88772904 | 6.78 |
ENSRNOT00000028265
|
Syne4l1
|
spectrin repeat containing, nuclear envelope family member 4-like 1 |
| chr3_+_111545007 | 6.64 |
ENSRNOT00000007247
|
Itpka
|
inositol-trisphosphate 3-kinase A |
| chr4_-_115354795 | 6.61 |
ENSRNOT00000017691
|
Cd207
|
CD207 molecule |
| chr6_+_132702448 | 5.95 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr8_-_111965889 | 5.85 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr7_-_135630654 | 5.79 |
ENSRNOT00000047388
ENSRNOT00000088223 ENSRNOT00000074793 |
Adamts20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr10_-_14937336 | 5.56 |
ENSRNOT00000025494
|
Sox8
|
SRY box 8 |
| chr12_+_22835019 | 5.52 |
ENSRNOT00000059530
|
Col26a1
|
collagen type XXVI alpha 1 chain |
| chr10_-_89900131 | 5.47 |
ENSRNOT00000028238
|
Sost
|
sclerostin |
| chr10_+_104019888 | 5.21 |
ENSRNOT00000032016
|
Slc16a5
|
solute carrier family 16 member 5 |
| chr8_-_48774898 | 5.18 |
ENSRNOT00000016303
|
Upk2
|
uroplakin 2 |
| chr17_+_82066152 | 5.11 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr8_+_116708027 | 5.05 |
ENSRNOT00000047309
|
Actl11
|
actin-like 11 |
| chr7_-_61798729 | 5.00 |
ENSRNOT00000010283
|
Dyrk2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr16_+_19800463 | 4.99 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr1_-_226152524 | 4.96 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr14_-_17436897 | 4.96 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr5_+_133620706 | 4.71 |
ENSRNOT00000053202
|
Gm12830
|
predicted gene 12830 |
| chr2_+_243728434 | 4.65 |
ENSRNOT00000017252
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr17_+_86199623 | 4.54 |
ENSRNOT00000022727
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr6_-_24563246 | 4.51 |
ENSRNOT00000074294
|
LOC685881
|
hypothetical protein LOC685881 |
| chr10_-_110585376 | 4.49 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr4_-_82160240 | 4.44 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
| chr3_+_8450275 | 4.43 |
ENSRNOT00000020073
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr4_-_82203154 | 4.39 |
ENSRNOT00000086210
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr8_+_50310405 | 4.28 |
ENSRNOT00000073507
|
Sik3
|
SIK family kinase 3 |
| chr5_-_74190991 | 4.27 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr18_-_40134504 | 4.27 |
ENSRNOT00000022294
|
Trim36
|
tripartite motif-containing 36 |
| chrX_+_53629255 | 4.24 |
ENSRNOT00000005021
|
Fthl17e
|
ferritin, heavy polypeptide-like 17, member E |
| chr4_+_159622404 | 4.20 |
ENSRNOT00000078299
|
Fgf23
|
fibroblast growth factor 23 |
| chr1_+_72635267 | 4.15 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr4_-_117575154 | 4.05 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr17_-_56992694 | 4.03 |
ENSRNOT00000079607
|
Cul2
|
cullin 2 |
| chr1_+_81230612 | 3.99 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr4_-_169093135 | 3.86 |
ENSRNOT00000011461
|
Pbp2
|
phosphatidylethanolamine binding protein 2 |
| chr1_+_101178104 | 3.85 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr10_-_56412544 | 3.79 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr10_-_94557764 | 3.78 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr7_-_143967484 | 3.76 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr9_-_81566642 | 3.75 |
ENSRNOT00000080345
|
Aamp
|
angio-associated, migratory cell protein |
| chr10_+_56237755 | 3.69 |
ENSRNOT00000016163
|
Fxr2
|
FMR1 autosomal homolog 2 |
| chr20_-_11737050 | 3.66 |
ENSRNOT00000001637
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr5_-_59198650 | 3.64 |
ENSRNOT00000020958
|
Olr833
|
olfactory receptor 833 |
| chr19_+_60017746 | 3.64 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr8_-_14373894 | 3.62 |
ENSRNOT00000011883
|
Mtnr1b
|
melatonin receptor 1B |
| chr3_-_94767239 | 3.46 |
ENSRNOT00000056808
|
Qser1
|
glutamine and serine rich 1 |
| chr18_-_61707307 | 3.46 |
ENSRNOT00000085784
ENSRNOT00000032047 |
Lman1
|
lectin, mannose-binding, 1 |
| chr5_-_159256699 | 3.43 |
ENSRNOT00000055867
|
Padi6
|
peptidyl arginine deiminase 6 |
| chr1_+_199248470 | 3.34 |
ENSRNOT00000025933
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr6_+_108032856 | 3.33 |
ENSRNOT00000014738
|
Zfp410
|
zinc finger protein 410 |
| chr8_-_53362006 | 3.33 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr13_+_51034256 | 3.29 |
ENSRNOT00000004528
ENSRNOT00000046854 ENSRNOT00000087320 |
Mybph
|
myosin binding protein H |
| chr2_+_203301838 | 3.25 |
ENSRNOT00000043146
|
Trim45
|
tripartite motif-containing 45 |
| chr2_+_199283909 | 3.13 |
ENSRNOT00000043194
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr12_+_25264192 | 3.12 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr3_-_10196626 | 3.12 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr15_+_30735921 | 3.08 |
ENSRNOT00000078887
|
AABR07017763.2
|
|
| chr5_-_137726333 | 3.08 |
ENSRNOT00000084932
|
Olr856
|
olfactory receptor 856 |
| chr10_+_46840113 | 3.07 |
ENSRNOT00000086083
ENSRNOT00000079133 |
Myo15a
|
myosin XVA |
| chr3_+_161212156 | 3.07 |
ENSRNOT00000020280
ENSRNOT00000083553 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr12_+_39913628 | 3.01 |
ENSRNOT00000046604
|
Ccdc63
|
coiled-coil domain containing 63 |
| chr15_-_29349800 | 2.98 |
ENSRNOT00000074218
|
RGD1563780
|
similar to RIKEN cDNA A430107P09 gene |
| chr16_-_74864816 | 2.96 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chrX_-_73360204 | 2.95 |
ENSRNOT00000091278
|
LOC100360296
|
BRCA2-interacting protein-like |
| chr1_-_104973648 | 2.95 |
ENSRNOT00000019739
|
Dbx1
|
developing brain homeobox 1 |
| chr10_-_85289777 | 2.91 |
ENSRNOT00000055427
ENSRNOT00000014863 |
Gpr179
|
G protein-coupled receptor 179 |
| chr3_-_81282157 | 2.86 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr1_-_88989552 | 2.85 |
ENSRNOT00000034001
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr5_-_58113553 | 2.84 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chr3_+_80833272 | 2.79 |
ENSRNOT00000023583
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr3_-_23474170 | 2.74 |
ENSRNOT00000039410
|
Scai
|
suppressor of cancer cell invasion |
| chr3_-_93734282 | 2.74 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr10_+_105787935 | 2.72 |
ENSRNOT00000000266
|
Mettl23
|
methyltransferase like 23 |
| chr2_+_27365148 | 2.69 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr9_+_41045363 | 2.63 |
ENSRNOT00000066284
|
Cfc1
|
cripto, FRL-1, cryptic family 1 |
| chr6_-_8066868 | 2.56 |
ENSRNOT00000008200
|
Lrpprc
|
leucine-rich pentatricopeptide repeat containing |
| chr19_-_11144641 | 2.53 |
ENSRNOT00000081246
|
Slc12a3
|
solute carrier family 12 member 3 |
| chr10_+_59923201 | 2.52 |
ENSRNOT00000073603
ENSRNOT00000045573 |
Olr1498
|
olfactory receptor 1498 |
| chrX_+_29157470 | 2.52 |
ENSRNOT00000081986
|
LOC100910245
|
ribose-phosphate pyrophosphokinase 2-like |
| chr16_+_81089292 | 2.50 |
ENSRNOT00000087192
ENSRNOT00000026150 |
Tfdp1
|
transcription factor Dp-1 |
| chr2_+_216863428 | 2.47 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr20_+_1936438 | 2.46 |
ENSRNOT00000073981
|
Olr1749
|
olfactory receptor 1749 |
| chr7_-_143966863 | 2.43 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr13_+_52887649 | 2.38 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr4_+_342302 | 2.38 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr7_-_143473697 | 2.35 |
ENSRNOT00000055336
ENSRNOT00000083027 |
Krt77
|
keratin 77 |
| chr8_+_71719563 | 2.33 |
ENSRNOT00000022828
|
Ppib
|
peptidylprolyl isomerase B |
| chr6_-_95502775 | 2.30 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr18_-_63488027 | 2.29 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr10_-_87541851 | 2.29 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chr20_+_4576514 | 2.28 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr7_-_140770647 | 2.23 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr1_+_123015746 | 2.21 |
ENSRNOT00000013483
|
Magel2
|
MAGE family member L2 |
| chr7_-_143217535 | 2.17 |
ENSRNOT00000088316
|
Kb15
|
type II keratin Kb15 |
| chr4_+_78024765 | 2.16 |
ENSRNOT00000043856
|
Krba1
|
KRAB-A domain containing 1 |
| chr3_-_176547965 | 2.15 |
ENSRNOT00000009041
|
Chrna4
|
cholinergic receptor nicotinic alpha 4 subunit |
| chr5_-_64622360 | 2.11 |
ENSRNOT00000073545
|
Olr841
|
olfactory receptor 841 |
| chr4_+_82665094 | 2.06 |
ENSRNOT00000078254
|
AABR07060591.1
|
|
| chr3_+_150055749 | 2.04 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr10_-_109622745 | 2.00 |
ENSRNOT00000074354
ENSRNOT00000088317 |
Pde6g
|
phosphodiesterase 6G |
| chr10_+_82937971 | 1.98 |
ENSRNOT00000005797
|
Dlx3
|
distal-less homeobox 3 |
| chr19_-_46101250 | 1.93 |
ENSRNOT00000015874
|
Adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr12_-_48218955 | 1.89 |
ENSRNOT00000067975
ENSRNOT00000080557 ENSRNOT00000000821 |
Acacb
|
acetyl-CoA carboxylase beta |
| chr14_+_60764409 | 1.89 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr5_+_162808646 | 1.84 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr10_-_39373437 | 1.79 |
ENSRNOT00000058907
|
Slc22a5
|
solute carrier family 22 member 5 |
| chr11_-_29618857 | 1.78 |
ENSRNOT00000064583
|
Krtap8-1
|
keratin associated protein 8-1 |
| chr9_+_99776886 | 1.77 |
ENSRNOT00000056602
|
Olr1352
|
olfactory receptor 1352 |
| chr10_-_12214822 | 1.76 |
ENSRNOT00000071005
|
Olr1365
|
olfactory receptor 1365 |
| chr5_-_157518511 | 1.74 |
ENSRNOT00000070865
ENSRNOT00000078028 |
Htr6
|
5-hydroxytryptamine receptor 6 |
| chr6_+_128750795 | 1.74 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr4_+_157513414 | 1.73 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr13_+_99220510 | 1.73 |
ENSRNOT00000004519
|
Tmem63a
|
transmembrane protein 63a |
| chr1_+_207654487 | 1.72 |
ENSRNOT00000025569
|
Foxi2
|
forkhead box I2 |
| chr20_+_5442390 | 1.72 |
ENSRNOT00000037499
|
Rps18
|
ribosomal protein S18 |
| chr3_+_150686638 | 1.71 |
ENSRNOT00000078235
|
Itch
|
itchy E3 ubiquitin protein ligase |
| chr7_-_120027026 | 1.70 |
ENSRNOT00000011215
|
Card10
|
caspase recruitment domain family, member 10 |
| chr18_-_25314047 | 1.68 |
ENSRNOT00000079778
|
AABR07031674.4
|
|
| chr1_+_207508414 | 1.64 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr5_+_162323373 | 1.64 |
ENSRNOT00000033150
|
Aadacl3
|
arylacetamide deacetylase-like 3 |
| chr3_-_151363115 | 1.64 |
ENSRNOT00000073492
|
Eif6
|
eukaryotic translation initiation factor 6 |
| chr6_-_99783047 | 1.63 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr1_+_282715344 | 1.63 |
ENSRNOT00000074399
|
LOC100910259
|
liver carboxylesterase-like |
| chr15_-_28835796 | 1.55 |
ENSRNOT00000060439
|
Olr1641
|
olfactory receptor 1641 |
| chr7_+_121311024 | 1.54 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr9_-_97151832 | 1.53 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr17_+_66446162 | 1.48 |
ENSRNOT00000078906
|
Heatr1
|
HEAT repeat containing 1 |
| chr3_-_73456572 | 1.45 |
ENSRNOT00000012872
|
Olr479
|
olfactory receptor 479 |
| chr18_-_28454756 | 1.44 |
ENSRNOT00000040091
|
Spata24
|
spermatogenesis associated 24 |
| chr4_+_113948514 | 1.42 |
ENSRNOT00000011521
|
LOC103692171
|
mannosyl-oligosaccharide glucosidase |
| chr8_-_36760742 | 1.42 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chrX_+_115561332 | 1.40 |
ENSRNOT00000076680
ENSRNOT00000075912 ENSRNOT00000007968 |
Alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr10_+_44292927 | 1.39 |
ENSRNOT00000058503
|
Olr1433
|
olfactory receptor 1433 |
| chr5_-_64615714 | 1.35 |
ENSRNOT00000007462
|
LOC500460
|
similar to olfactory receptor Olr841 |
| chr4_-_180887050 | 1.33 |
ENSRNOT00000065730
|
Ints13
|
integrator complex subunit 13 |
| chr5_-_107388182 | 1.33 |
ENSRNOT00000058358
|
LOC100912356
|
interferon alpha-1-like |
| chr8_-_55681930 | 1.32 |
ENSRNOT00000065744
|
Colca2
|
colorectal cancer associated 2 |
| chr1_+_101669086 | 1.29 |
ENSRNOT00000028524
|
Ntn5
|
netrin 5 |
| chr7_-_4293352 | 1.27 |
ENSRNOT00000047489
|
Olr984
|
olfactory receptor 984 |
| chr20_-_47306318 | 1.24 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr8_-_97115027 | 1.18 |
ENSRNOT00000018685
|
Ankrd34c
|
ankyrin repeat domain 34C |
| chr5_+_140979435 | 1.17 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr17_+_66446569 | 1.17 |
ENSRNOT00000070825
|
Heatr1
|
HEAT repeat containing 1 |
| chr5_+_160825419 | 1.15 |
ENSRNOT00000080134
|
AABR07050317.1
|
|
| chr1_-_89045586 | 1.10 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr1_-_170916059 | 1.10 |
ENSRNOT00000041974
|
Olr217
|
olfactory receptor 217 |
| chr2_-_119140110 | 1.09 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr10_-_61045355 | 1.08 |
ENSRNOT00000048167
|
Olr1514
|
olfactory receptor 1514 |
| chr3_-_77792014 | 0.98 |
ENSRNOT00000091085
|
Olr671
|
olfactory receptor 671 |
| chrX_+_28435507 | 0.97 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr2_+_78407414 | 0.97 |
ENSRNOT00000084541
ENSRNOT00000087109 |
Zfp622
|
zinc finger protein 622 |
| chr12_-_39881352 | 0.95 |
ENSRNOT00000083296
|
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr17_+_45801528 | 0.95 |
ENSRNOT00000089221
|
Olr1664
|
olfactory receptor 1664 |
| chr6_+_135156984 | 0.89 |
ENSRNOT00000009709
ENSRNOT00000082595 |
Wdr20
|
WD repeat domain 20 |
| chr1_+_260093641 | 0.88 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr10_+_36477692 | 0.88 |
ENSRNOT00000044464
|
Olr1405
|
olfactory receptor 1405 |
| chr7_-_120026755 | 0.86 |
ENSRNOT00000077677
|
Card10
|
caspase recruitment domain family, member 10 |
| chr5_+_106952082 | 0.84 |
ENSRNOT00000046931
|
RGD1561246
|
similar to put. precursor MuIFN-alpha 5 |
| chr1_+_220746387 | 0.83 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr7_+_41115154 | 0.82 |
ENSRNOT00000066174
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr1_+_170796271 | 0.81 |
ENSRNOT00000042820
|
Olr210
|
olfactory receptor 210 |
| chr5_+_68717519 | 0.80 |
ENSRNOT00000066226
|
Smc2
|
structural maintenance of chromosomes 2 |
| chr4_+_6931495 | 0.79 |
ENSRNOT00000079770
|
Wdr86
|
WD repeat domain 86 |
| chr1_-_219640294 | 0.78 |
ENSRNOT00000054850
|
Kdm2a
|
lysine demethylase 2A |
| chr10_-_12571316 | 0.75 |
ENSRNOT00000074440
|
Olr1372
|
olfactory receptor 1372 |
| chr3_-_8981362 | 0.74 |
ENSRNOT00000086717
|
Crat
|
carnitine O-acetyltransferase |
| chr7_+_121480723 | 0.73 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr4_-_67520356 | 0.71 |
ENSRNOT00000014604
|
Braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr3_+_175885894 | 0.68 |
ENSRNOT00000039780
|
Ntsr1
|
neurotensin receptor 1 |
| chr2_-_206997519 | 0.66 |
ENSRNOT00000027042
|
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr1_+_220423426 | 0.66 |
ENSRNOT00000072647
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr20_+_9948908 | 0.60 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr10_-_35833861 | 0.58 |
ENSRNOT00000049942
|
Canx
|
calnexin |
| chr19_-_10138159 | 0.56 |
ENSRNOT00000018082
|
Tepp
|
testis, prostate and placenta expressed |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 43.2 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 2.8 | 8.5 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 2.4 | 78.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 2.0 | 16.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 2.0 | 9.9 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.9 | 7.7 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 1.8 | 5.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.5 | 6.0 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 1.4 | 4.2 | GO:1904383 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) response to sodium phosphate(GO:1904383) |
| 1.4 | 5.6 | GO:0072034 | retinal rod cell differentiation(GO:0060221) renal vesicle induction(GO:0072034) |
| 1.2 | 3.6 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 1.2 | 11.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.0 | 3.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 1.0 | 5.0 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.0 | 23.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.9 | 18.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.8 | 10.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.8 | 2.3 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.8 | 4.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.8 | 2.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) regulation of prolactin signaling pathway(GO:1902211) regulation of interleukin-4-mediated signaling pathway(GO:1902214) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.8 | 2.3 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.8 | 4.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.7 | 3.4 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.7 | 8.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.7 | 2.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.7 | 2.6 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.6 | 5.8 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.6 | 3.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 1.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.6 | 1.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.6 | 8.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.6 | 2.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.5 | 2.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.5 | 3.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.5 | 2.6 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.5 | 5.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.5 | 1.9 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.5 | 3.8 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 3.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.4 | 4.3 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.4 | 1.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 1.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 5.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.4 | 3.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.4 | 4.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.4 | 2.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 6.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 2.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 4.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.3 | 3.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.3 | 1.6 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.3 | 4.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 6.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.3 | 5.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.3 | 3.9 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.3 | 2.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 13.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.3 | 3.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 2.6 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 2.0 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.7 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 1.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.7 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.2 | 7.2 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 2.9 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 8.2 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.2 | 1.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.2 | 3.7 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.2 | 0.6 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.2 | 0.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 3.8 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901028) |
| 0.2 | 3.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 2.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 0.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.9 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.2 | 2.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 2.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 1.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.7 | GO:2000301 | negative regulation of fibroblast migration(GO:0010764) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.8 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 3.1 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 0.8 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 4.3 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 1.4 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 1.3 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 12.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.7 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 2.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 4.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.4 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.1 | 22.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 4.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 2.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 4.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 4.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.6 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 3.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.6 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 5.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 4.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 3.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 2.5 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.0 | 0.3 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 4.4 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.2 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 23.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0045136 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.2 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.7 | GO:0042254 | ribosome biogenesis(GO:0042254) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0031309 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
| 1.2 | 8.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.8 | 2.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.8 | 2.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.8 | 9.9 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.7 | 8.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 3.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.5 | 13.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.4 | 3.5 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.4 | 4.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 6.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.3 | 1.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 4.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 5.5 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.3 | 3.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 0.7 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 6.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 2.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 5.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.2 | 11.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 1.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 3.5 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 0.1 | 8.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 2.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 2.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 14.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 5.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 9.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 3.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 16.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 115.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 4.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 6.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 2.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 18.6 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 30.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 4.3 | 13.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 2.0 | 23.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.9 | 7.7 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 1.9 | 7.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.6 | 4.7 | GO:0051903 | all-trans retinal binding(GO:0005503) benzaldehyde dehydrogenase activity(GO:0019115) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 1.2 | 5.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 1.0 | 8.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.0 | 3.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.8 | 4.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.8 | 2.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.7 | 6.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 3.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.7 | 3.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.7 | 78.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.6 | 1.8 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) antibiotic transporter activity(GO:0042895) |
| 0.6 | 4.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.6 | 10.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.6 | 2.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 3.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.5 | 2.6 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.5 | 13.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.5 | 6.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.5 | 27.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.5 | 1.9 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 3.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.4 | 2.7 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 3.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 3.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 6.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 2.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.3 | 2.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.3 | 5.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 3.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 4.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 22.9 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.2 | 0.8 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 3.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 3.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.7 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.2 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 16.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 2.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.1 | 1.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 3.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 4.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 5.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 2.0 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 1.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 5.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 7.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 3.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 5.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.3 | GO:0070063 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) RNA polymerase binding(GO:0070063) |
| 0.1 | 1.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 4.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.9 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 3.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 17.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 3.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 24.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 8.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 13.8 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 2.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.8 | 18.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.6 | 8.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 11.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.4 | 93.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 14.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 2.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 7.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 4.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 5.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 10.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 8.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 3.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 16.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.3 | 6.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 9.9 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.4 | 14.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 4.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 3.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 9.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 8.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 42.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 2.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 1.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 4.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 4.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 2.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.7 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 5.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 3.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |