Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
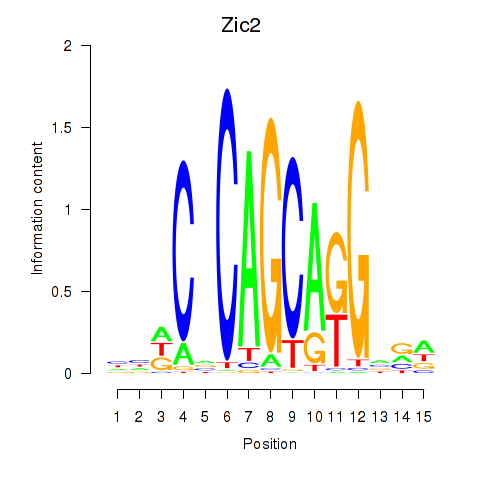
Results for Zic2
Z-value: 0.95
Transcription factors associated with Zic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic2
|
ENSRNOG00000054879 | Zic family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic2 | rn6_v1_chr15_+_108908607_108908607 | -0.11 | 5.2e-02 | Click! |
Activity profile of Zic2 motif
Sorted Z-values of Zic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_57855416 | 20.40 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chr6_+_139551751 | 20.16 |
ENSRNOT00000081684
|
AABR07065699.2
|
|
| chr8_-_49308806 | 19.82 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr5_-_160403373 | 17.73 |
ENSRNOT00000018393
|
Ctrc
|
chymotrypsin C |
| chr1_+_279867034 | 15.74 |
ENSRNOT00000024164
|
Pnliprp1
|
pancreatic lipase-related protein 1 |
| chr4_-_69268336 | 15.51 |
ENSRNOT00000018042
|
Prss3b
|
protease, serine, 3B |
| chr3_+_17107861 | 15.22 |
ENSRNOT00000043097
|
AABR07051563.1
|
|
| chr6_-_139719323 | 15.01 |
ENSRNOT00000090133
|
AABR07065705.4
|
|
| chr3_+_16817051 | 14.84 |
ENSRNOT00000071666
|
AABR07051550.1
|
|
| chr14_-_100217913 | 14.79 |
ENSRNOT00000079167
|
Plek
|
pleckstrin |
| chr6_-_138772736 | 14.60 |
ENSRNOT00000071492
|
AABR07065651.1
|
|
| chr6_-_138772894 | 14.16 |
ENSRNOT00000080779
|
AABR07065651.1
|
|
| chr5_+_154522119 | 14.16 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr8_+_55603968 | 12.89 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr6_-_140216072 | 12.45 |
ENSRNOT00000072365
|
AABR07065750.1
|
|
| chr17_-_34905117 | 12.36 |
ENSRNOT00000088931
|
Irf4
|
interferon regulatory factor 4 |
| chr19_-_37907714 | 12.28 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr1_-_215858034 | 11.49 |
ENSRNOT00000027656
|
Ins2
|
insulin 2 |
| chr1_-_226791773 | 11.09 |
ENSRNOT00000082482
ENSRNOT00000065376 ENSRNOT00000054812 ENSRNOT00000086669 |
LOC100911215
|
T-cell surface glycoprotein CD5-like |
| chr6_-_45669148 | 10.94 |
ENSRNOT00000010092
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr1_-_89543967 | 10.81 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr17_-_42640221 | 10.71 |
ENSRNOT00000018699
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr7_-_12741296 | 10.45 |
ENSRNOT00000060648
|
Arhgap45
|
Rho GTPase activating protein 45 |
| chr10_-_56506446 | 10.21 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr17_-_15881327 | 10.05 |
ENSRNOT00000022341
|
Ninj1
|
ninjurin 1 |
| chr10_+_103438303 | 9.97 |
ENSRNOT00000085262
|
Cd300a
|
Cd300a molecule |
| chr17_-_43504604 | 9.83 |
ENSRNOT00000083829
ENSRNOT00000066313 |
Slc17a1
|
solute carrier family 17 member 1 |
| chr14_+_77067503 | 9.67 |
ENSRNOT00000085275
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr9_-_52912293 | 9.59 |
ENSRNOT00000005228
|
Slc40a1
|
solute carrier family 40 member 1 |
| chr2_-_203680083 | 9.53 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr2_-_123282214 | 9.26 |
ENSRNOT00000021156
|
Ccna2
|
cyclin A2 |
| chr6_-_138744480 | 9.15 |
ENSRNOT00000089387
|
AABR07065651.5
|
|
| chr10_-_15577977 | 8.82 |
ENSRNOT00000052292
|
Hba-a3
|
hemoglobin alpha, adult chain 3 |
| chr6_+_129052503 | 8.68 |
ENSRNOT00000044152
|
RGD1565462
|
similar to BC049975 protein |
| chr2_-_123281856 | 8.50 |
ENSRNOT00000079745
|
Ccna2
|
cyclin A2 |
| chrX_+_1311121 | 8.35 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr6_-_140418831 | 8.31 |
ENSRNOT00000086301
|
AABR07065768.2
|
|
| chr15_-_29446332 | 8.23 |
ENSRNOT00000082901
|
AABR07017634.1
|
|
| chrX_+_15273933 | 8.16 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr1_-_261051498 | 8.11 |
ENSRNOT00000071417
|
Arhgap19
|
Rho GTPase activating protein 19 |
| chr17_-_32783427 | 8.04 |
ENSRNOT00000059921
|
Serpinb6b
|
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| chr6_-_139747579 | 7.95 |
ENSRNOT00000075502
|
AABR07065705.3
|
|
| chr6_+_128048099 | 7.95 |
ENSRNOT00000084685
ENSRNOT00000087017 |
LOC500712
|
Ab1-233 |
| chr2_+_209838869 | 7.87 |
ENSRNOT00000092365
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr15_+_32817343 | 7.78 |
ENSRNOT00000073853
|
AABR07017902.1
|
|
| chr15_-_29987717 | 7.72 |
ENSRNOT00000071501
|
AABR07017676.2
|
|
| chr6_-_138507294 | 7.64 |
ENSRNOT00000078516
|
AABR07065640.1
|
|
| chr12_-_52452040 | 7.49 |
ENSRNOT00000067453
|
Pole
|
DNA polymerase epsilon, catalytic subunit |
| chr9_+_16530074 | 7.48 |
ENSRNOT00000091565
|
Ptcra
|
pre T-cell antigen receptor alpha |
| chrX_-_156891038 | 7.42 |
ENSRNOT00000091495
|
Avpr2
|
arginine vasopressin receptor 2 |
| chr10_+_44271587 | 7.35 |
ENSRNOT00000047700
|
Trim58
|
tripartite motif-containing 58 |
| chr6_-_99783047 | 7.32 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr6_+_127927650 | 7.31 |
ENSRNOT00000057271
|
LOC100909605
|
serine protease inhibitor A3F-like |
| chr6_-_132608600 | 7.31 |
ENSRNOT00000015855
|
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr10_+_57040267 | 7.29 |
ENSRNOT00000026207
|
Arrb2
|
arrestin, beta 2 |
| chr10_-_90307658 | 7.26 |
ENSRNOT00000092102
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr8_+_33514042 | 7.08 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr5_-_160620334 | 7.06 |
ENSRNOT00000019171
|
Tmem51
|
transmembrane protein 51 |
| chr20_-_11620945 | 7.04 |
ENSRNOT00000079725
|
Krtap12-2
|
keratin associated protein 12-2 |
| chr20_+_4020317 | 7.01 |
ENSRNOT00000000526
|
RT1-DOb
|
RT1 class II, locus DOb |
| chr8_+_71822129 | 7.00 |
ENSRNOT00000089147
|
Dapk2
|
death-associated protein kinase 2 |
| chr3_+_16590244 | 6.99 |
ENSRNOT00000073229
|
AABR07051535.1
|
|
| chr16_+_74886719 | 6.98 |
ENSRNOT00000089265
|
Atp7b
|
ATPase copper transporting beta |
| chr9_+_95285592 | 6.96 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr16_+_20109200 | 6.94 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr7_-_78847974 | 6.93 |
ENSRNOT00000006004
|
Dpys
|
dihydropyrimidinase |
| chr3_+_18706988 | 6.86 |
ENSRNOT00000074650
|
AABR07051652.1
|
|
| chr11_+_71211768 | 6.86 |
ENSRNOT00000079753
|
LOC100910650
|
uncharacterized LOC100910650 |
| chr16_+_19800463 | 6.69 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr10_+_15697227 | 6.54 |
ENSRNOT00000028008
|
Il9r
|
interleukin 9 receptor |
| chrX_+_70461718 | 6.49 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr1_+_141986145 | 6.48 |
ENSRNOT00000030914
|
Sema4b
|
semaphorin 4B |
| chr14_-_5378726 | 6.46 |
ENSRNOT00000002896
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr6_-_138507135 | 6.46 |
ENSRNOT00000071078
|
AABR07065640.1
|
|
| chr1_-_204605552 | 6.44 |
ENSRNOT00000022904
|
Nkx1-2
|
NK1 homeobox 2 |
| chr16_-_7026540 | 6.43 |
ENSRNOT00000051751
|
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr10_-_65692016 | 6.40 |
ENSRNOT00000085074
ENSRNOT00000038690 |
Slc13a2
|
solute carrier family 13 member 2 |
| chr15_-_28104206 | 6.35 |
ENSRNOT00000032536
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr6_+_139523495 | 6.31 |
ENSRNOT00000075467
|
AABR07065699.4
|
|
| chr20_+_4852671 | 6.21 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr4_-_122237754 | 6.11 |
ENSRNOT00000029915
|
Chst13
|
carbohydrate sulfotransferase 13 |
| chr1_-_48360131 | 6.06 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr1_+_192233910 | 6.06 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr17_+_43734461 | 6.01 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chr1_-_227932603 | 6.01 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr2_-_202816562 | 6.00 |
ENSRNOT00000020401
|
Fam46c
|
family with sequence similarity 46, member C |
| chr10_-_87067456 | 5.96 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr11_+_45462345 | 5.92 |
ENSRNOT00000029420
|
Nit2
|
nitrilase family, member 2 |
| chr3_-_5975734 | 5.91 |
ENSRNOT00000081376
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr10_-_56935141 | 5.84 |
ENSRNOT00000025910
|
Alox12e
|
arachidonate 12-lipoxygenase, epidermal |
| chr11_-_60882379 | 5.81 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr6_-_138536162 | 5.77 |
ENSRNOT00000083031
|
AABR07065643.1
|
|
| chr11_+_54619129 | 5.73 |
ENSRNOT00000059924
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chrX_-_135342996 | 5.70 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr2_-_219090874 | 5.62 |
ENSRNOT00000019377
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr7_-_12634641 | 5.54 |
ENSRNOT00000093132
|
Cfd
|
complement factor D |
| chr1_+_31967978 | 5.53 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr6_+_144291974 | 5.52 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr3_-_2727616 | 5.50 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr1_-_254671596 | 5.47 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr8_+_91368430 | 5.44 |
ENSRNOT00000051612
|
Ttk
|
Ttk protein kinase |
| chr7_+_126939925 | 5.44 |
ENSRNOT00000022746
|
Gramd4
|
GRAM domain containing 4 |
| chr17_-_9791781 | 5.41 |
ENSRNOT00000090536
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr20_+_4852496 | 5.34 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr1_-_164101578 | 5.27 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr6_+_123974798 | 5.25 |
ENSRNOT00000075794
|
Kcnk13
|
potassium two pore domain channel subfamily K member 13 |
| chr1_-_88920291 | 5.23 |
ENSRNOT00000028306
|
Kirrel2
|
kin of IRRE like 2 (Drosophila) |
| chr13_-_41738622 | 5.17 |
ENSRNOT00000004520
ENSRNOT00000084552 |
Actr3
|
ARP3 actin related protein 3 homolog |
| chr5_+_34040258 | 5.15 |
ENSRNOT00000009758
|
Ggh
|
gamma-glutamyl hydrolase |
| chr16_-_81434038 | 5.11 |
ENSRNOT00000067508
|
Rasa3
|
RAS p21 protein activator 3 |
| chr13_+_82438697 | 5.10 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr15_+_31436302 | 5.08 |
ENSRNOT00000071012
|
AABR07017825.2
|
|
| chr8_-_66893083 | 5.06 |
ENSRNOT00000037028
ENSRNOT00000091755 |
Kif23
|
kinesin family member 23 |
| chr15_+_45422010 | 5.04 |
ENSRNOT00000012231
|
Rnaseh2b
|
ribonuclease H2, subunit B |
| chr16_-_48692476 | 4.97 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr9_+_98505259 | 4.90 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr5_+_157801163 | 4.88 |
ENSRNOT00000024160
|
Akr7a3
|
aldo-keto reductase family 7 member A3 |
| chr7_-_12635943 | 4.85 |
ENSRNOT00000015029
|
Cfd
|
complement factor D |
| chr6_-_51498337 | 4.79 |
ENSRNOT00000012487
|
Pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr4_+_96252798 | 4.78 |
ENSRNOT00000008947
|
RGD1560028
|
similar to RIKEN cDNA C130060K24 gene |
| chr16_+_9023387 | 4.77 |
ENSRNOT00000067761
|
RGD1561145
|
similar to novel protein |
| chr7_-_12899004 | 4.77 |
ENSRNOT00000011086
|
Gzmm
|
granzyme M |
| chr7_-_12646960 | 4.77 |
ENSRNOT00000014687
|
Prtn3
|
proteinase 3 |
| chr12_+_30198822 | 4.74 |
ENSRNOT00000041645
|
Gusb
|
glucuronidase, beta |
| chr15_-_27733785 | 4.71 |
ENSRNOT00000029488
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr3_-_5976244 | 4.67 |
ENSRNOT00000009893
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr11_-_70499200 | 4.66 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr3_+_2591331 | 4.65 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr10_+_18996523 | 4.64 |
ENSRNOT00000046135
|
Lcp2
|
lymphocyte cytosolic protein 2 |
| chr1_+_21748261 | 4.57 |
ENSRNOT00000019519
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr10_-_82374171 | 4.56 |
ENSRNOT00000032693
|
Eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr17_-_9792007 | 4.55 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr10_-_70337532 | 4.53 |
ENSRNOT00000055963
|
Slfn13
|
schlafen family member 13 |
| chr10_+_59529785 | 4.51 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr8_-_49573742 | 4.46 |
ENSRNOT00000021926
|
Il10ra
|
interleukin 10 receptor subunit alpha |
| chr7_-_130128589 | 4.45 |
ENSRNOT00000079777
ENSRNOT00000009325 |
Mapk11
|
mitogen-activated protein kinase 11 |
| chr15_-_29976615 | 4.45 |
ENSRNOT00000060396
|
AABR07017676.1
|
|
| chr15_-_44627765 | 4.45 |
ENSRNOT00000058887
|
Dock5
|
dedicator of cytokinesis 5 |
| chr2_+_240396152 | 4.44 |
ENSRNOT00000034565
|
Cenpe
|
centromere protein E |
| chr12_-_17322608 | 4.43 |
ENSRNOT00000033038
|
LOC102546864
|
uncharacterized LOC102546864 |
| chr3_+_81283137 | 4.42 |
ENSRNOT00000008986
ENSRNOT00000086217 |
Pex16
|
peroxisomal biogenesis factor 16 |
| chr1_-_48559162 | 4.34 |
ENSRNOT00000080352
|
Plg
|
plasminogen |
| chr9_+_10956056 | 4.32 |
ENSRNOT00000073930
|
Plin5
|
perilipin 5 |
| chr5_-_155204456 | 4.30 |
ENSRNOT00000089574
|
Ephb2
|
Eph receptor B2 |
| chr8_+_91368111 | 4.30 |
ENSRNOT00000083151
|
Ttk
|
Ttk protein kinase |
| chrX_-_72078551 | 4.27 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr18_+_12056113 | 4.27 |
ENSRNOT00000038450
|
Dsg4
|
desmoglein 4 |
| chr2_-_104830898 | 4.23 |
ENSRNOT00000050440
|
Hps3
|
Hermansky-Pudlak syndrome 3 |
| chr11_-_61499557 | 4.23 |
ENSRNOT00000046208
|
Usf3
|
upstream transcription factor family member 3 |
| chr16_+_74752655 | 4.21 |
ENSRNOT00000029266
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr3_-_176744377 | 4.21 |
ENSRNOT00000017787
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr3_+_159368273 | 4.21 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr8_-_6061837 | 4.14 |
ENSRNOT00000007702
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr7_+_141258584 | 4.11 |
ENSRNOT00000083570
|
Aqp6
|
aquaporin 6 |
| chr2_+_263895241 | 4.11 |
ENSRNOT00000014126
ENSRNOT00000014034 |
Ptger3
|
prostaglandin E receptor 3 |
| chr20_+_40778927 | 4.08 |
ENSRNOT00000001081
|
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr2_+_209839299 | 4.06 |
ENSRNOT00000092450
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr8_-_120446455 | 4.03 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr12_-_22478752 | 4.00 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr5_-_169630340 | 4.00 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr11_-_32508420 | 3.99 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr3_+_5624506 | 3.98 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr1_-_254671778 | 3.98 |
ENSRNOT00000025493
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr10_-_87510629 | 3.96 |
ENSRNOT00000084197
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr4_+_62220736 | 3.91 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr10_-_87535438 | 3.90 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr11_+_47260641 | 3.88 |
ENSRNOT00000078856
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr16_-_48981980 | 3.88 |
ENSRNOT00000014235
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chrX_-_23187341 | 3.87 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr1_+_282567674 | 3.85 |
ENSRNOT00000090543
|
Ces2i
|
carboxylesterase 2I |
| chr10_+_86950557 | 3.83 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr14_+_17228856 | 3.81 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr10_-_57739109 | 3.79 |
ENSRNOT00000082171
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr19_+_15139920 | 3.76 |
ENSRNOT00000074479
ENSRNOT00000020775 |
Ces1f
|
carboxylesterase 1F |
| chr5_+_126668689 | 3.76 |
ENSRNOT00000036072
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr3_+_161236898 | 3.72 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr12_-_41625403 | 3.71 |
ENSRNOT00000001876
|
Sds
|
serine dehydratase |
| chr15_+_31164401 | 3.71 |
ENSRNOT00000084286
|
AABR07017798.1
|
|
| chr7_-_3342491 | 3.71 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr8_+_48382121 | 3.70 |
ENSRNOT00000008685
|
Thy1
|
Thy-1 cell surface antigen |
| chr12_+_52452273 | 3.69 |
ENSRNOT00000056680
ENSRNOT00000088381 |
Pxmp2
|
peroxisomal membrane protein 2 |
| chr10_+_4313100 | 3.68 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr1_+_221596148 | 3.66 |
ENSRNOT00000028536
|
Gpha2
|
glycoprotein hormone alpha 2 |
| chr10_-_87261717 | 3.64 |
ENSRNOT00000015740
|
Krt27
|
keratin 27 |
| chr19_-_9843673 | 3.63 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr13_-_100450209 | 3.63 |
ENSRNOT00000090712
|
Lbr
|
lamin B receptor |
| chr8_+_59507990 | 3.62 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr8_+_5768811 | 3.62 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr14_-_81472952 | 3.61 |
ENSRNOT00000058111
|
Sh3bp2
|
SH3-domain binding protein 2 |
| chr10_-_64915148 | 3.55 |
ENSRNOT00000048543
|
Sez6
|
seizure related 6 homolog |
| chr5_-_136053210 | 3.53 |
ENSRNOT00000025903
|
Kif2c
|
kinesin family member 2C |
| chr10_-_2229493 | 3.51 |
ENSRNOT00000047732
|
LOC688899
|
similar to 40S ribosomal protein S19 |
| chr7_-_123892429 | 3.50 |
ENSRNOT00000037681
|
Nfam1
|
NFAT activating protein with ITAM motif 1 |
| chr6_-_127337791 | 3.48 |
ENSRNOT00000032015
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr2_-_30577218 | 3.48 |
ENSRNOT00000024674
|
Ocln
|
occludin |
| chr1_-_256813711 | 3.48 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr5_-_68139199 | 3.48 |
ENSRNOT00000009987
|
Plppr1
|
phospholipid phosphatase related 1 |
| chr6_-_129010271 | 3.47 |
ENSRNOT00000075378
|
Serpina10
|
serpin family A member 10 |
| chr1_+_83573032 | 3.46 |
ENSRNOT00000083650
|
AC142154.2
|
|
| chr8_-_75995371 | 3.45 |
ENSRNOT00000014014
|
Foxb1
|
forkhead box B1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 3.9 | 11.6 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 3.8 | 11.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 3.7 | 14.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 3.6 | 10.9 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 3.6 | 10.8 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 3.3 | 10.0 | GO:0060101 | negative regulation of phagocytosis, engulfment(GO:0060101) negative regulation of eosinophil activation(GO:1902567) negative regulation of eosinophil migration(GO:2000417) |
| 3.3 | 10.0 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 3.3 | 19.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 3.2 | 9.7 | GO:1905132 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 3.2 | 9.5 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 3.1 | 18.5 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 2.4 | 7.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 2.4 | 9.6 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 2.4 | 7.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 2.3 | 7.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 2.3 | 7.0 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 2.3 | 7.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 2.3 | 6.9 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 2.3 | 9.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 2.2 | 8.7 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 2.1 | 10.7 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 2.1 | 14.8 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 2.0 | 8.2 | GO:0030221 | basophil differentiation(GO:0030221) |
| 2.0 | 6.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 2.0 | 11.9 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 2.0 | 9.9 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 1.9 | 7.5 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.8 | 7.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 1.7 | 10.5 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.7 | 5.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) regulation of bleb assembly(GO:1904170) |
| 1.6 | 4.9 | GO:0009407 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 1.5 | 4.6 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 1.5 | 4.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.5 | 7.3 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 1.4 | 7.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 1.4 | 4.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.4 | 4.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) positive regulation of actin filament-based movement(GO:1903116) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 1.3 | 6.7 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.3 | 5.3 | GO:0097680 | maintenance of Golgi location(GO:0051684) double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 1.3 | 6.4 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 1.2 | 3.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.2 | 3.5 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 1.1 | 4.6 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.1 | 5.7 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 1.1 | 3.3 | GO:0010979 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) calcitriol biosynthetic process from calciol(GO:0036378) vitamin D catabolic process(GO:0042369) positive regulation of response to alcohol(GO:1901421) |
| 1.1 | 4.3 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 1.0 | 19.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 1.0 | 3.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 1.0 | 3.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.0 | 3.0 | GO:0046968 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 1.0 | 5.9 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 1.0 | 2.9 | GO:0009946 | inhibition of neuroepithelial cell differentiation(GO:0002085) proximal/distal axis specification(GO:0009946) |
| 1.0 | 2.9 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 1.0 | 3.9 | GO:1901423 | response to benzene(GO:1901423) |
| 1.0 | 9.5 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.0 | 2.9 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.9 | 3.7 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.9 | 3.7 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.9 | 4.5 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.9 | 4.4 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.9 | 2.6 | GO:0046356 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.9 | 3.5 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.9 | 6.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.8 | 5.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.8 | 3.4 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.8 | 7.0 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 10.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.8 | 6.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.8 | 6.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.7 | 4.4 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.7 | 2.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.7 | 0.7 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.7 | 5.7 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 7.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 5.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.7 | 2.8 | GO:1904253 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.7 | 7.5 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.7 | 2.0 | GO:0006083 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 0.7 | 2.0 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.6 | 2.6 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.6 | 2.6 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.6 | 3.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 2.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 1.8 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.6 | 1.8 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.6 | 8.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.6 | 1.8 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.6 | 5.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.6 | 4.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.6 | 2.9 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.6 | 3.4 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.6 | 3.4 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.5 | 2.6 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.5 | 2.6 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.5 | 2.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.5 | 6.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 3.5 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 | 4.0 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.5 | 4.0 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.5 | 1.5 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.5 | 1.5 | GO:0006431 | methionyl-tRNA aminoacylation(GO:0006431) |
| 0.5 | 3.4 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.5 | 3.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.5 | 3.4 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.5 | 2.4 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.5 | 4.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.5 | 4.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.4 | 4.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.4 | 3.9 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.4 | 4.1 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.4 | 5.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.4 | 1.2 | GO:1904760 | myofibroblast differentiation(GO:0036446) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) regulation of myofibroblast differentiation(GO:1904760) |
| 0.4 | 2.4 | GO:0033504 | floor plate development(GO:0033504) |
| 0.4 | 10.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.4 | 3.5 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.4 | 2.6 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.4 | 1.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.4 | 2.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.4 | 10.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.4 | 1.8 | GO:1902732 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.4 | 1.8 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 12.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.3 | 1.0 | GO:0035793 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling(GO:0060683) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) embryonic lung development(GO:1990401) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.3 | 1.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 1.0 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.3 | 2.4 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.3 | 1.0 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.3 | 1.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 | 1.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.3 | 3.3 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.3 | 2.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 0.9 | GO:1903796 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 0.3 | 1.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 1.2 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.3 | 2.8 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 0.6 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.3 | 2.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 2.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 0.9 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.3 | 5.1 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.3 | 1.7 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.3 | 5.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 5.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 2.5 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.3 | 3.8 | GO:0050930 | positive regulation of cAMP-mediated signaling(GO:0043950) induction of positive chemotaxis(GO:0050930) |
| 0.3 | 3.8 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.3 | 3.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 4.4 | GO:1902358 | sulfate transport(GO:0008272) sulfate transmembrane transport(GO:1902358) |
| 0.3 | 7.7 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.3 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 0.5 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.2 | 5.1 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 1.4 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.2 | 1.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.2 | 4.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 1.8 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 1.5 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 8.9 | GO:0006778 | porphyrin-containing compound metabolic process(GO:0006778) |
| 0.2 | 0.8 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 | 1.5 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 3.4 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 0.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.7 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.2 | 1.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 1.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 0.7 | GO:0097116 | postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.2 | 0.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 1.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 1.5 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.2 | 6.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 2.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 2.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 2.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.3 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 2.4 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 | 1.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 2.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.9 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 2.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 4.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 3.0 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.4 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) positive regulation of glucagon secretion(GO:0070094) sensory neuron migration(GO:1904937) |
| 0.1 | 3.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 0.4 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.1 | 1.9 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.1 | 11.8 | GO:0031016 | pancreas development(GO:0031016) |
| 0.1 | 3.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 4.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 1.0 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.1 | 1.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 8.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 1.9 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 1.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.3 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 6.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.4 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 1.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 7.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 9.4 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.1 | 0.7 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.7 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.4 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.9 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 5.0 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 2.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 0.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 7.5 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 4.2 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.9 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 1.2 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 4.2 | GO:0043473 | pigmentation(GO:0043473) |
| 0.1 | 12.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.8 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 3.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 3.3 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 4.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.6 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 8.6 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 1.7 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 1.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 3.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 2.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 1.4 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.8 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 2.5 | 19.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.2 | 8.7 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.6 | 4.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.5 | 7.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.4 | 5.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.3 | 5.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.3 | 5.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.3 | 5.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.2 | 6.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 1.2 | 4.6 | GO:0036398 | TCR signalosome(GO:0036398) |
| 1.1 | 4.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.0 | 15.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 1.0 | 5.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 3.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.8 | 4.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.8 | 8.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.8 | 59.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.8 | 2.4 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.8 | 3.8 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.7 | 7.3 | GO:0008091 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 0.7 | 7.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.7 | 5.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.7 | 18.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 2.6 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.6 | 2.9 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.6 | 5.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.5 | 1.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.5 | 10.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.5 | 3.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) dendritic spine neck(GO:0044326) |
| 0.5 | 1.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.5 | 13.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.5 | 11.8 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.4 | 7.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 5.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 8.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.4 | 3.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 1.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.4 | 2.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.4 | 5.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 4.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.3 | 5.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 2.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 4.2 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.3 | 1.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.3 | 3.3 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 7.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 10.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 4.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 22.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 4.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.2 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 3.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 1.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 3.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 2.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 5.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 8.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 10.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 3.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 9.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 11.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 5.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 5.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 7.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 12.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.0 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 4.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 2.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.4 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 8.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 55.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 4.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 3.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 7.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 2.4 | 7.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 2.4 | 7.3 | GO:0031826 | follicle-stimulating hormone receptor binding(GO:0031762) type 2A serotonin receptor binding(GO:0031826) |
| 2.0 | 6.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 2.0 | 6.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.8 | 7.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.7 | 6.9 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 1.6 | 8.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.6 | 4.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.6 | 14.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.5 | 6.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 1.5 | 4.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 1.5 | 5.9 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.5 | 5.8 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 1.4 | 4.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 1.4 | 7.0 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.3 | 4.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.3 | 2.6 | GO:0015116 | sulfate transmembrane transporter activity(GO:0015116) |
| 1.3 | 5.1 | GO:0042806 | fucose binding(GO:0042806) |
| 1.2 | 3.7 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 1.2 | 3.7 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 1.2 | 6.0 | GO:0032564 | dATP binding(GO:0032564) |
| 1.1 | 9.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 1.0 | 5.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.0 | 10.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.0 | 3.0 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.0 | 9.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.0 | 2.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 1.0 | 9.7 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 10.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.9 | 4.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.9 | 2.6 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.8 | 21.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.8 | 2.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.8 | 3.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.7 | 8.7 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.7 | 4.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.7 | 4.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.7 | 7.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.7 | 9.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.7 | 1.3 | GO:0038100 | nodal binding(GO:0038100) |
| 0.7 | 3.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 13.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.7 | 7.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.7 | 2.6 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.6 | 5.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.6 | 3.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.6 | 14.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.6 | 3.7 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.6 | 2.4 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.6 | 1.8 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.6 | 7.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.6 | 6.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.6 | 8.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.6 | 4.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.6 | 1.8 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.6 | 4.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.6 | 3.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.6 | 4.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.6 | 5.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.6 | 1.7 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.5 | 11.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.5 | 4.9 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 3.0 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.5 | 1.5 | GO:0004825 | methionine-tRNA ligase activity(GO:0004825) |
| 0.5 | 2.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 3.8 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 5.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.5 | 14.5 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.5 | 1.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.5 | 9.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.5 | 3.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.4 | 9.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.4 | 6.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 6.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 4.9 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 4.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.4 | 1.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.4 | 7.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 1.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 1.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 4.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 4.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 3.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 1.9 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.4 | 2.6 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.4 | 4.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.4 | 2.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 4.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.4 | 1.8 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 3.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 17.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 13.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 2.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 3.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 11.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.3 | 5.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 1.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 63.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 0.9 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.3 | 2.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 31.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 11.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 0.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 5.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.3 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 3.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 12.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 4.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 1.5 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 2.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 6.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 3.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 7.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 3.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 13.7 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 4.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 3.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 2.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 1.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 8.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 4.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 5.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 14.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 1.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.4 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 2.8 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 1.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 6.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 2.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 6.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0086077 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.7 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 0.1 | 2.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 0.9 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 4.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 4.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 5.2 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 3.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 4.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 2.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 5.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 6.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 5.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 9.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 2.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.8 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 0.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 5.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 5.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 5.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 4.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.6 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 4.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.6 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 1.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 28.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.8 | 10.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.6 | 10.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.6 | 20.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.6 | 6.9 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.6 | 8.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.5 | 22.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 32.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 8.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 5.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 6.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 12.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 4.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 12.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 15.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 5.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 4.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 4.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 2.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 19.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.2 | 18.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.9 | 7.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.7 | 9.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.7 | 6.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.6 | 4.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.5 | 6.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 7.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 6.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 10.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 8.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 6.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 26.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.5 | 7.7 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.5 | 5.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 7.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.4 | 4.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.4 | 11.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 4.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 3.4 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.4 | 5.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 7.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 9.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 3.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 8.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 7.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 7.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 5.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.3 | 29.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 3.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 6.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 2.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 11.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 4.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 10.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 4.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 7.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 7.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 6.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 2.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 3.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 6.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 8.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 9.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 3.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 8.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 8.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 2.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.8 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 0.1 | 0.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 0.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.8 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 3.6 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 3.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 4.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |