Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
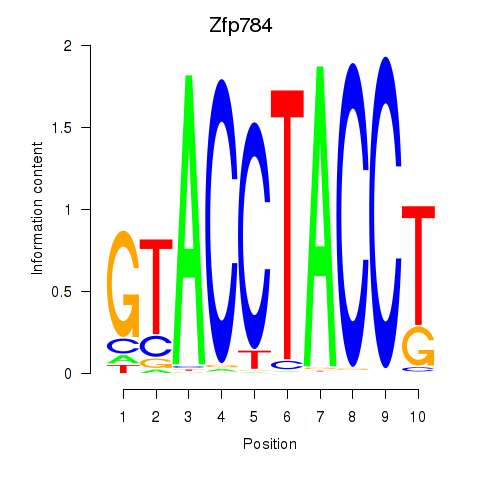
Results for Zfp784
Z-value: 1.01
Transcription factors associated with Zfp784
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp865 | rn6_v1_chr1_+_72356880_72356880 | 0.52 | 1.4e-23 | Click! |
Activity profile of Zfp784 motif
Sorted Z-values of Zfp784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_64554953 | 25.99 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr5_+_49311030 | 23.28 |
ENSRNOT00000010850
|
Cnr1
|
cannabinoid receptor 1 |
| chr20_-_31956649 | 21.67 |
ENSRNOT00000072429
|
Hk1
|
hexokinase 1 |
| chr15_+_15275541 | 20.50 |
ENSRNOT00000012153
|
Cadps
|
calcium dependent secretion activator |
| chr3_-_153893847 | 18.23 |
ENSRNOT00000085938
|
LOC100911769
|
band 4.1-like protein 1-like |
| chr7_+_142912316 | 17.94 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_-_113937941 | 17.68 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr16_-_20097287 | 17.05 |
ENSRNOT00000025162
|
Unc13a
|
unc-13 homolog A |
| chr13_-_1946508 | 16.93 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr2_-_200625434 | 16.73 |
ENSRNOT00000079754
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr7_+_117723263 | 16.66 |
ENSRNOT00000085152
|
Kifc2
|
kinesin family member C2 |
| chr1_-_48825364 | 16.60 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr8_-_49038169 | 16.41 |
ENSRNOT00000047303
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr3_+_38559914 | 16.36 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr14_-_43072843 | 16.31 |
ENSRNOT00000064263
|
Limch1
|
LIM and calponin homology domains 1 |
| chr7_-_139223116 | 16.07 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr8_-_116361343 | 15.97 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr1_+_168964202 | 15.66 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr3_+_152552822 | 15.61 |
ENSRNOT00000089719
|
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr15_-_52317219 | 15.61 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr1_-_168972725 | 15.25 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr3_+_14467330 | 15.21 |
ENSRNOT00000078939
|
Gsn
|
gelsolin |
| chr2_-_210454737 | 15.11 |
ENSRNOT00000079993
|
Ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr2_-_200692335 | 14.93 |
ENSRNOT00000026290
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr1_+_29225707 | 14.83 |
ENSRNOT00000018849
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr3_-_164055561 | 14.60 |
ENSRNOT00000064849
|
B4galt5
|
beta-1,4-galactosyltransferase 5 |
| chr4_-_56114254 | 14.38 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr10_+_107455845 | 14.27 |
ENSRNOT00000004373
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr6_-_42616548 | 14.12 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr11_-_11585078 | 14.08 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr8_+_118333706 | 14.03 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr6_-_27190126 | 13.96 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr12_+_25036605 | 13.88 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr1_+_168945449 | 13.60 |
ENSRNOT00000087661
ENSRNOT00000019913 |
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr19_+_53723822 | 13.51 |
ENSRNOT00000023478
|
RGD1304884
|
similar to RIKEN cDNA 6430548M08 |
| chr1_-_81881549 | 12.95 |
ENSRNOT00000027497
|
Atp1a3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr11_+_84396033 | 12.80 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr2_+_199199845 | 12.74 |
ENSRNOT00000020932
|
LOC100909441
|
tubulin alpha-1C chain-like |
| chr11_-_24641820 | 12.58 |
ENSRNOT00000044081
ENSRNOT00000048854 |
App
|
amyloid beta precursor protein |
| chr10_-_89130339 | 12.46 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr17_+_11101306 | 12.36 |
ENSRNOT00000030893
|
Drd1
|
dopamine receptor D1 |
| chr8_-_130127392 | 12.12 |
ENSRNOT00000026159
|
Cck
|
cholecystokinin |
| chr10_+_23661343 | 12.06 |
ENSRNOT00000047970
|
Ebf1
|
early B-cell factor 1 |
| chr8_-_128754514 | 12.04 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr12_-_39850567 | 11.76 |
ENSRNOT00000001712
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr3_+_172374957 | 11.33 |
ENSRNOT00000035916
|
Gnas
|
GNAS complex locus |
| chr5_+_74727494 | 11.32 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chrX_-_156379189 | 11.26 |
ENSRNOT00000083148
|
Plxna3
|
plexin A3 |
| chr19_+_25043680 | 11.25 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr3_-_29996865 | 11.22 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr7_+_11582984 | 11.16 |
ENSRNOT00000026893
|
Gng7
|
G protein subunit gamma 7 |
| chr13_-_49313940 | 10.59 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chr19_+_54553419 | 10.10 |
ENSRNOT00000025392
|
Jph3
|
junctophilin 3 |
| chr9_-_19372673 | 10.00 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr1_+_171793782 | 9.81 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr16_+_74810938 | 9.77 |
ENSRNOT00000058091
|
Nek5
|
NIMA-related kinase 5 |
| chr7_-_140617721 | 9.47 |
ENSRNOT00000081355
|
Tuba1b
|
tubulin, alpha 1B |
| chr10_-_56831243 | 9.34 |
ENSRNOT00000025403
|
Slc16a13
|
solute carrier family 16, member 13 |
| chr2_-_231409988 | 9.33 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr3_+_145032200 | 9.32 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr4_-_157252104 | 9.21 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chrX_-_43592200 | 9.17 |
ENSRNOT00000005033
|
Acot9
|
acyl-CoA thioesterase 9 |
| chrX_-_71168612 | 9.17 |
ENSRNOT00000075934
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr3_+_110442637 | 9.03 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr8_+_111600532 | 9.03 |
ENSRNOT00000081952
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr10_+_84167331 | 9.02 |
ENSRNOT00000010965
|
Hoxb4
|
homeo box B4 |
| chr1_+_219403970 | 8.70 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr20_-_4132604 | 8.52 |
ENSRNOT00000077630
ENSRNOT00000048332 |
RT1-Da
|
RT1 class II, locus Da |
| chrX_+_78196300 | 8.43 |
ENSRNOT00000048695
|
P2ry10
|
purinergic receptor P2Y10 |
| chr10_-_64657089 | 8.38 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr6_+_9483594 | 8.34 |
ENSRNOT00000089272
|
AABR07062799.2
|
|
| chr12_+_52359310 | 8.05 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr5_-_148392689 | 7.70 |
ENSRNOT00000018464
ENSRNOT00000080166 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr5_+_118574801 | 7.65 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr4_+_56805132 | 7.47 |
ENSRNOT00000010001
ENSRNOT00000085483 |
Irf5
|
interferon regulatory factor 5 |
| chr9_+_43889473 | 7.33 |
ENSRNOT00000024330
|
Inpp4a
|
inositol polyphosphate-4-phosphatase type I A |
| chr6_-_39363367 | 7.14 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr8_+_132241134 | 6.89 |
ENSRNOT00000006055
|
Clec3b
|
C-type lectin domain family 3, member B |
| chrX_-_31851715 | 6.78 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chr14_-_45165207 | 6.74 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr8_-_21968415 | 6.71 |
ENSRNOT00000067325
ENSRNOT00000064932 |
Dnmt1
|
DNA methyltransferase 1 |
| chr2_-_231648122 | 6.67 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr5_+_155660553 | 6.66 |
ENSRNOT00000081893
|
Wnt4
|
wingless-type MMTV integration site family, member 4 |
| chr13_-_26768708 | 6.59 |
ENSRNOT00000092763
|
Bcl2
|
BCL2, apoptosis regulator |
| chr1_+_222672501 | 6.58 |
ENSRNOT00000051855
|
RGD1560108
|
similar to RIKEN cDNA 2700081O15 |
| chr10_+_23661013 | 6.37 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr2_+_72006099 | 6.22 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr10_+_84182118 | 5.87 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr4_+_88328061 | 5.77 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr9_+_116652530 | 5.73 |
ENSRNOT00000029210
|
L3mbtl4
|
l(3)mbt-like 4 (Drosophila) |
| chr2_-_241545250 | 5.70 |
ENSRNOT00000073875
|
Bank1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr5_+_145079803 | 5.52 |
ENSRNOT00000084202
|
Sfpq
|
splicing factor proline and glutamine rich |
| chr1_+_69615603 | 5.51 |
ENSRNOT00000084806
|
Zfp418
|
zinc finger protein 418 |
| chr1_+_276659542 | 5.46 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr20_+_46168177 | 5.39 |
ENSRNOT00000057175
|
Zbtb24
|
zinc finger and BTB domain containing 24 |
| chr7_-_144837583 | 5.35 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr2_+_257568613 | 5.33 |
ENSRNOT00000066125
|
Usp33
|
ubiquitin specific peptidase 33 |
| chr4_+_7377563 | 5.24 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr4_+_155709613 | 5.24 |
ENSRNOT00000013272
|
Necap1
|
NECAP endocytosis associated 1 |
| chr18_-_37096132 | 5.19 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_69615434 | 4.96 |
ENSRNOT00000020751
|
Zfp418
|
zinc finger protein 418 |
| chr12_+_32103198 | 4.90 |
ENSRNOT00000085464
|
Tmem132d
|
transmembrane protein 132D |
| chr3_+_56861396 | 4.84 |
ENSRNOT00000000008
ENSRNOT00000084375 |
Gad1
|
glutamate decarboxylase 1 |
| chr4_-_119889949 | 4.83 |
ENSRNOT00000033687
|
H1fx
|
H1 histone family, member X |
| chr1_-_82166363 | 4.80 |
ENSRNOT00000027774
|
Pafah1b3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 |
| chr1_-_259674425 | 4.73 |
ENSRNOT00000091116
|
LOC108348083
|
delta-1-pyrroline-5-carboxylate synthase |
| chr1_-_259089632 | 4.67 |
ENSRNOT00000020940
ENSRNOT00000091297 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr3_+_148541909 | 4.64 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr14_-_84189266 | 4.57 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr5_+_147714163 | 4.57 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr15_-_24199341 | 4.57 |
ENSRNOT00000015553
|
Dlgap5
|
DLG associated protein 5 |
| chr2_-_46042636 | 4.54 |
ENSRNOT00000087161
|
AABR07008152.1
|
|
| chr7_-_140502441 | 4.31 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chr17_+_81455955 | 4.28 |
ENSRNOT00000044313
|
Slc39a12
|
solute carrier family 39 member 12 |
| chr1_-_88826302 | 4.26 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chrX_+_123404518 | 4.17 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr7_-_124367630 | 4.12 |
ENSRNOT00000055978
|
Ttll1
|
tubulin tyrosine ligase like 1 |
| chr8_-_49308806 | 4.08 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr1_+_84685931 | 4.02 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr1_+_280383579 | 3.99 |
ENSRNOT00000064022
ENSRNOT00000042977 |
Kcnk18
|
potassium two pore domain channel subfamily K member 18 |
| chr9_+_100489852 | 3.91 |
ENSRNOT00000022854
|
Ppp1r7
|
protein phosphatase 1, regulatory subunit 7 |
| chr9_-_71852113 | 3.89 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr6_-_91518996 | 3.81 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr8_+_127144903 | 3.76 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chr1_+_225184939 | 3.68 |
ENSRNOT00000079456
|
Ahnak
|
AHNAK nucleoprotein |
| chr11_-_14304603 | 3.67 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr17_-_77527894 | 3.66 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr3_-_9822182 | 3.55 |
ENSRNOT00000076637
ENSRNOT00000010115 |
RGD1305178
|
similar to Hypothetical protein MGC11690 |
| chr1_-_16203838 | 3.53 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr9_-_20195566 | 3.52 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr12_+_22153983 | 3.42 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr3_-_10226286 | 3.40 |
ENSRNOT00000093627
|
Fubp3
|
far upstream element binding protein 3 |
| chr2_-_211001258 | 3.39 |
ENSRNOT00000037336
|
Atxn7l2
|
ataxin 7-like 2 |
| chr7_-_124286478 | 3.39 |
ENSRNOT00000088125
|
Pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_-_58198782 | 3.32 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr1_+_1784078 | 3.31 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr2_-_184263564 | 3.28 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr17_-_56909992 | 3.23 |
ENSRNOT00000021801
|
Bambi
|
BMP and activin membrane-bound inhibitor |
| chr3_+_91191837 | 3.18 |
ENSRNOT00000006097
|
Rag2
|
recombination activating 2 |
| chr7_+_122979021 | 3.18 |
ENSRNOT00000085707
|
Zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr7_+_145068286 | 3.14 |
ENSRNOT00000088956
ENSRNOT00000065753 |
Nckap1l
|
NCK associated protein 1 like |
| chr4_+_88832178 | 3.11 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr2_-_205212681 | 3.05 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chrX_-_13116743 | 3.00 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr5_-_164844586 | 2.99 |
ENSRNOT00000011287
|
Clcn6
|
chloride voltage-gated channel 6 |
| chrX_+_48196916 | 2.94 |
ENSRNOT00000045976
|
Da2-19
|
uncharacterized LOC302729 |
| chr3_-_170378210 | 2.91 |
ENSRNOT00000055102
|
Aurka
|
aurora kinase A |
| chr9_-_100306829 | 2.87 |
ENSRNOT00000038563
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr18_+_72550219 | 2.85 |
ENSRNOT00000046847
|
Smad2
|
SMAD family member 2 |
| chr4_+_157513414 | 2.82 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr14_-_5006594 | 2.82 |
ENSRNOT00000076571
|
Zfp326
|
zinc finger protein 326 |
| chr17_+_6684621 | 2.81 |
ENSRNOT00000089138
|
RGD1311345
|
similar to CG9752-PA |
| chr10_-_62032400 | 2.80 |
ENSRNOT00000086378
ENSRNOT00000004161 |
Dph1
|
diphthamide biosynthesis 1 |
| chr14_+_106153575 | 2.76 |
ENSRNOT00000010264
|
Vps54
|
VPS54 GARP complex subunit |
| chr10_+_103703404 | 2.73 |
ENSRNOT00000086469
|
Rab37
|
RAB37, member RAS oncogene family |
| chr16_+_50111306 | 2.70 |
ENSRNOT00000019302
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr14_+_100373129 | 2.69 |
ENSRNOT00000029980
|
Wdr92
|
WD repeat domain 92 |
| chr8_-_131899023 | 2.65 |
ENSRNOT00000034690
|
Zfp445
|
zinc finger protein 445 |
| chr5_+_172488708 | 2.49 |
ENSRNOT00000019930
|
Morn1
|
MORN repeat containing 1 |
| chr1_-_72464492 | 2.43 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr10_-_13075864 | 2.39 |
ENSRNOT00000005220
|
Paqr4
|
progestin and adipoQ receptor family member 4 |
| chr11_-_68197772 | 2.35 |
ENSRNOT00000003060
ENSRNOT00000081875 |
Hspbap1
|
Hspb associated protein 1 |
| chr6_-_86924146 | 2.22 |
ENSRNOT00000029411
|
Mis18bp1
|
MIS18 binding protein 1 |
| chr5_+_107711077 | 2.22 |
ENSRNOT00000029486
|
Mtap
|
methylthioadenosine phosphorylase |
| chr1_-_234670113 | 2.20 |
ENSRNOT00000017133
|
LOC499331
|
similar to hypothetical protein D030056L22 |
| chr16_+_32449116 | 2.19 |
ENSRNOT00000050549
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr10_+_52334240 | 2.16 |
ENSRNOT00000005518
|
Zfp18
|
zinc finger protein 18 |
| chr5_+_152325871 | 2.07 |
ENSRNOT00000044711
|
Ftl1
|
ferritin light chain 1 |
| chr9_-_100306194 | 2.05 |
ENSRNOT00000087584
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr6_+_50528823 | 2.00 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr8_-_94922017 | 2.00 |
ENSRNOT00000083934
|
Cep162
|
centrosomal protein 162 |
| chr13_+_101790865 | 1.98 |
ENSRNOT00000087784
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I subunit A |
| chr11_-_66759402 | 1.77 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chrX_+_105575759 | 1.76 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr15_+_28413461 | 1.72 |
ENSRNOT00000060521
|
Tmem253
|
transmembrane protein 253 |
| chr12_-_38274036 | 1.71 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr19_+_33928356 | 1.63 |
ENSRNOT00000058162
|
Ednra
|
endothelin receptor type A |
| chr1_-_72339395 | 1.63 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr7_+_24939498 | 1.59 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr17_+_84881414 | 1.55 |
ENSRNOT00000034157
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr13_-_50261438 | 1.45 |
ENSRNOT00000051824
|
LOC498231
|
LRRGT00144 |
| chr6_-_129509720 | 1.35 |
ENSRNOT00000006131
|
Atg2b
|
autophagy related 2B |
| chr1_-_101449829 | 1.35 |
ENSRNOT00000028315
|
LOC100360087
|
ferritin light chain 1-like |
| chr5_+_78483893 | 1.33 |
ENSRNOT00000059183
ENSRNOT00000059181 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr7_-_144837395 | 1.29 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr1_+_80777014 | 1.27 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr10_+_104523996 | 1.27 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr12_-_39881352 | 1.20 |
ENSRNOT00000083296
|
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr12_+_15857805 | 1.20 |
ENSRNOT00000001667
|
Iqce
|
IQ motif containing E |
| chr7_+_2458833 | 1.20 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr1_+_165237847 | 1.14 |
ENSRNOT00000022963
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr11_-_33003021 | 1.14 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr1_-_221917901 | 1.11 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr6_-_99843245 | 1.02 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr1_+_47942800 | 0.99 |
ENSRNOT00000079312
|
Wtap
|
Wilms tumor 1 associated protein |
| chr7_-_116063078 | 0.99 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
| chr1_-_257498844 | 0.95 |
ENSRNOT00000019056
|
Noc3l
|
NOC3-like DNA replication regulator |
| chr12_+_38274297 | 0.95 |
ENSRNOT00000087905
ENSRNOT00000057788 |
Rsrc2
|
arginine and serine rich coiled-coil 2 |
| chr6_+_80108655 | 0.89 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr16_+_81756971 | 0.88 |
ENSRNOT00000044085
|
Pcid2
|
PCI domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp784
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 5.4 | 37.6 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 5.1 | 15.2 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 4.0 | 16.0 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 3.9 | 15.6 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 3.8 | 15.2 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 3.8 | 11.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 3.7 | 11.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 3.6 | 21.7 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 3.6 | 14.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 3.5 | 10.6 | GO:0060167 | clustering of voltage-gated potassium channels(GO:0045163) regulation of adenosine receptor signaling pathway(GO:0060167) |
| 3.4 | 16.9 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 3.3 | 26.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 3.3 | 6.7 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 3.1 | 12.4 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 3.0 | 12.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 3.0 | 17.9 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 2.8 | 8.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 2.3 | 9.2 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 2.2 | 6.6 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) cellular response to fluoride(GO:1902618) |
| 2.2 | 15.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 2.1 | 29.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.0 | 31.7 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 2.0 | 5.9 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 1.8 | 16.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 1.8 | 25.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 1.8 | 5.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 1.7 | 5.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.7 | 6.7 | GO:0090309 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.6 | 11.3 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.5 | 4.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 1.5 | 14.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 1.4 | 53.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 1.4 | 13.0 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 1.4 | 8.5 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC protein complex(GO:0002501) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.3 | 9.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.3 | 16.4 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 1.2 | 12.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.1 | 3.3 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 1.1 | 3.3 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 1.1 | 3.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.0 | 3.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 1.0 | 4.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.0 | 12.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 1.0 | 6.8 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.0 | 4.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.0 | 2.9 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte maturation(GO:1900195) |
| 0.9 | 9.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.9 | 9.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.9 | 3.7 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.9 | 5.5 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.9 | 9.2 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.9 | 4.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.9 | 14.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.9 | 4.3 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.8 | 11.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.8 | 3.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.8 | 5.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.8 | 3.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.7 | 2.2 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.7 | 2.9 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.7 | 10.0 | GO:0060088 | diet induced thermogenesis(GO:0002024) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.7 | 8.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.7 | 2.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.6 | 6.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.6 | 7.5 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.6 | 1.8 | GO:0042531 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.6 | 13.9 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.6 | 2.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.5 | 2.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.5 | 1.0 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.5 | 10.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.5 | 3.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.5 | 5.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.5 | 4.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.5 | 7.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.4 | 14.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.4 | 4.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 3.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.4 | 4.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 1.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) positive regulation of progesterone secretion(GO:2000872) |
| 0.4 | 3.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.4 | 5.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.3 | 1.6 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.3 | 2.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 4.0 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.3 | 0.9 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 3.2 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.3 | 3.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 9.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 9.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 15.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 13.5 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.2 | 5.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) protein K48-linked deubiquitination(GO:0071108) |
| 0.2 | 3.0 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 | 7.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.2 | 1.3 | GO:0061450 | trophoblast cell migration(GO:0061450) |
| 0.1 | 3.7 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) |
| 0.1 | 9.8 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.1 | 0.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 1.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 3.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 0.6 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 15.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 3.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 3.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 2.8 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 2.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 14.9 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.6 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 4.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 6.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.8 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 11.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.6 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.7 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 5.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 7.7 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.9 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 3.4 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 10.8 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 3.1 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 5.2 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 2.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 4.3 | GO:0007409 | axonogenesis(GO:0007409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 4.2 | 25.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 4.2 | 12.6 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 4.0 | 44.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.8 | 14.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 2.5 | 15.2 | GO:0030478 | actin cap(GO:0030478) |
| 2.1 | 17.1 | GO:0044305 | calyx of Held(GO:0044305) presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.9 | 7.7 | GO:0033503 | HULC complex(GO:0033503) |
| 1.9 | 15.0 | GO:0043203 | axon hillock(GO:0043203) |
| 1.8 | 5.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 1.7 | 13.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.3 | 20.5 | GO:0031045 | dense core granule(GO:0031045) |
| 1.1 | 10.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.0 | 16.4 | GO:0045180 | basal cortex(GO:0045180) |
| 1.0 | 6.9 | GO:0001652 | granular component(GO:0001652) |
| 0.9 | 2.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.9 | 3.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.8 | 5.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 3.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.7 | 4.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.7 | 2.8 | GO:0000938 | GARP complex(GO:0000938) |
| 0.7 | 10.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.7 | 3.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.6 | 12.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.6 | 1.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 22.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.5 | 8.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 14.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 2.0 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 0.5 | 16.0 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 13.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.4 | 37.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 5.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.4 | 13.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 2.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.4 | 3.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 4.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 11.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 2.0 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.3 | 4.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 19.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 14.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.3 | 7.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 0.9 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.3 | 16.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 2.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 10.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 5.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 14.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 9.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 38.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 0.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 3.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 20.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 5.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 5.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 5.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 36.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 1.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 8.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 4.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 5.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 9.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 5.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.9 | GO:0043679 | axon terminus(GO:0043679) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 23.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 5.6 | 16.9 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 5.3 | 31.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 5.1 | 15.2 | GO:0031720 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 4.1 | 12.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 3.8 | 15.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 3.8 | 15.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 3.6 | 21.7 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 3.1 | 12.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 2.9 | 29.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.8 | 14.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 2.4 | 7.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 2.4 | 12.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 2.4 | 11.8 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 2.3 | 9.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 1.9 | 17.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 1.9 | 11.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.8 | 12.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.6 | 14.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.6 | 9.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.5 | 9.2 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 1.4 | 16.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 1.3 | 6.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 1.2 | 4.8 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.2 | 14.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.1 | 6.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 1.1 | 18.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 1.0 | 9.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.0 | 23.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.0 | 4.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.9 | 3.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.9 | 13.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.8 | 6.8 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.8 | 11.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.7 | 5.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.7 | 2.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.7 | 2.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.7 | 17.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.7 | 8.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.7 | 31.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.7 | 3.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 5.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.6 | 11.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 4.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.5 | 10.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.5 | 4.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 8.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 13.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 1.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 11.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 9.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 3.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.3 | 1.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 23.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.3 | 3.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 3.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 4.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 23.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 11.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 2.0 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 17.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 16.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 14.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 2.9 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 3.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 3.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 16.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 10.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 4.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 9.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 5.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 10.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 2.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 3.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 10.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 3.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 13.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 7.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 5.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 3.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 3.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 7.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 7.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 5.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 3.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 3.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 6.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.4 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 1.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 6.9 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 2.7 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 4.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 22.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.8 | 38.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.5 | 17.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 26.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 9.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 17.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.3 | 24.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 53.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 15.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 9.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.3 | 3.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 10.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 9.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.2 | 8.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 4.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 14.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 4.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 6.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 11.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 35.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.5 | 2.9 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.1 | 12.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.1 | 25.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.9 | 6.6 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.9 | 22.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.9 | 13.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.9 | 14.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.7 | 8.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.7 | 12.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.6 | 9.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 4.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.5 | 14.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 9.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 16.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.5 | 17.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.5 | 15.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.4 | 19.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 21.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.3 | 3.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 14.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 12.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 15.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 3.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 4.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 10.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 3.8 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.2 | 6.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 17.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 4.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 3.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 7.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 18.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 8.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 15.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 3.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 3.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 4.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |