Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
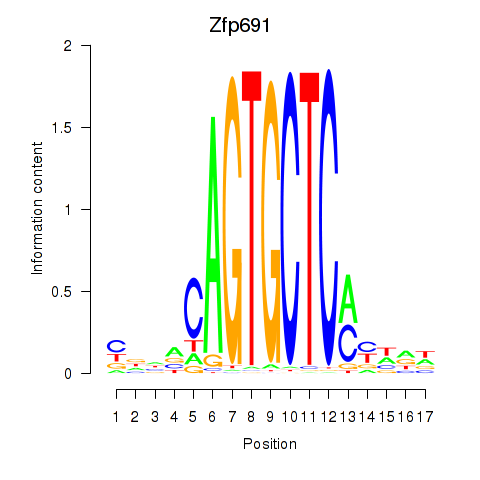
Results for Zfp691
Z-value: 1.33
Transcription factors associated with Zfp691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp691
|
ENSRNOG00000007398 | zinc finger protein 691 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp691 | rn6_v1_chr5_-_138222534_138222534 | -0.34 | 4.2e-10 | Click! |
Activity profile of Zfp691 motif
Sorted Z-values of Zfp691 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_32811135 | 61.37 |
ENSRNOT00000067689
|
AABR07017902.1
|
|
| chr11_-_37914983 | 54.47 |
ENSRNOT00000039876
|
Mx1
|
myxovirus (influenza virus) resistance 1 |
| chr10_-_110232843 | 46.72 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr14_-_15258207 | 46.23 |
ENSRNOT00000039383
|
Cxcl13
|
C-X-C motif chemokine ligand 13 |
| chr13_+_50164563 | 45.85 |
ENSRNOT00000029533
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr6_-_138662365 | 44.89 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr6_+_139486942 | 42.66 |
ENSRNOT00000080320
|
AABR07065699.3
|
|
| chr6_-_138900915 | 40.33 |
ENSRNOT00000075363
|
AABR07065656.3
|
|
| chr6_+_139486775 | 39.95 |
ENSRNOT00000077771
|
AABR07065699.3
|
|
| chr10_-_31359699 | 37.88 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_-_49308806 | 37.78 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chrX_-_71169038 | 36.24 |
ENSRNOT00000005343
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr6_+_139560028 | 35.45 |
ENSRNOT00000072633
|
LOC100360581
|
rCG58847-like |
| chr6_+_139428999 | 34.47 |
ENSRNOT00000084482
|
AABR07065693.2
|
|
| chrX_-_71168612 | 32.98 |
ENSRNOT00000075934
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr11_+_71872830 | 32.60 |
ENSRNOT00000050364
|
Nrros
|
negative regulator of reactive oxygen species |
| chr13_-_91981432 | 31.35 |
ENSRNOT00000004637
|
AABR07021804.1
|
|
| chr7_+_125288081 | 31.25 |
ENSRNOT00000085216
|
Parvg
|
parvin, gamma |
| chr15_-_29548400 | 31.18 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr6_+_139293130 | 30.98 |
ENSRNOT00000071714
|
LOC100359993
|
Ighg protein-like |
| chr6_-_138931262 | 30.92 |
ENSRNOT00000078126
ENSRNOT00000042118 |
LOC100359993
|
Ighg protein-like |
| chr6_-_139004980 | 28.14 |
ENSRNOT00000085427
ENSRNOT00000087351 |
AABR07065656.4
|
|
| chr8_+_133210473 | 27.41 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr6_-_139660666 | 26.44 |
ENSRNOT00000086098
|
AABR07065705.1
|
|
| chr6_-_139710905 | 26.37 |
ENSRNOT00000077430
|
AABR07065705.2
|
|
| chr6_-_138954741 | 24.80 |
ENSRNOT00000083278
|
AABR07065656.7
|
|
| chr6_+_139293294 | 24.14 |
ENSRNOT00000050297
ENSRNOT00000081817 |
LOC100359993
|
Ighg protein-like |
| chr6_-_138954577 | 24.08 |
ENSRNOT00000042728
|
AABR07065656.7
|
|
| chr8_-_22270647 | 24.02 |
ENSRNOT00000028380
|
S1pr5
|
sphingosine-1-phosphate receptor 5 |
| chr10_-_90415070 | 23.54 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr10_-_67401836 | 22.98 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr11_+_60383431 | 22.77 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr13_-_26768708 | 22.23 |
ENSRNOT00000092763
|
Bcl2
|
BCL2, apoptosis regulator |
| chr1_-_133559975 | 21.12 |
ENSRNOT00000046645
|
Mctp2
|
multiple C2 and transmembrane domain containing 2 |
| chr20_-_32232632 | 21.04 |
ENSRNOT00000068184
|
Ddx21
|
DExD-box helicase 21 |
| chrX_+_1311121 | 20.86 |
ENSRNOT00000038909
|
Cfp
|
complement factor properdin |
| chr1_+_215628785 | 20.44 |
ENSRNOT00000054864
|
Lsp1
|
lymphocyte-specific protein 1 |
| chr10_-_13619935 | 20.06 |
ENSRNOT00000064392
|
Ccnf
|
cyclin F |
| chr16_-_21362955 | 19.72 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chr20_-_32139789 | 19.45 |
ENSRNOT00000078140
|
Srgn
|
serglycin |
| chr14_-_14426437 | 19.15 |
ENSRNOT00000068134
|
Anxa3
|
annexin A3 |
| chr8_+_132869712 | 18.89 |
ENSRNOT00000008294
|
Cxcr6
|
C-X-C motif chemokine receptor 6 |
| chr6_-_139637187 | 18.66 |
ENSRNOT00000089454
|
LOC100359993
|
Ighg protein-like |
| chr10_-_14613878 | 18.56 |
ENSRNOT00000024072
|
Baiap3
|
BAI1-associated protein 3 |
| chr3_-_7202848 | 18.17 |
ENSRNOT00000078428
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr1_-_13050644 | 18.17 |
ENSRNOT00000081259
|
Heca
|
hdc homolog, cell cycle regulator |
| chr3_-_7203420 | 17.87 |
ENSRNOT00000015236
|
Gfi1b
|
growth factor independent 1B transcriptional repressor |
| chr3_+_2591331 | 17.74 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr7_-_60860990 | 17.54 |
ENSRNOT00000009511
|
Rap1b
|
RAP1B, member of RAS oncogene family |
| chr6_-_139637354 | 16.85 |
ENSRNOT00000072900
|
LOC100359993
|
Ighg protein-like |
| chr1_+_256955652 | 15.90 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr14_+_5928737 | 15.59 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr19_+_37600148 | 14.96 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr15_-_108326907 | 14.90 |
ENSRNOT00000016848
|
Gpr18
|
G protein-coupled receptor 18 |
| chr19_-_9843673 | 14.82 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr6_-_138931429 | 14.72 |
ENSRNOT00000090584
|
LOC100359993
|
Ighg protein-like |
| chr1_-_227505267 | 13.53 |
ENSRNOT00000028443
|
Ms4a7
|
membrane spanning 4-domains A7 |
| chr8_+_113105814 | 13.28 |
ENSRNOT00000016749
|
Cpne4
|
copine 4 |
| chr5_+_47894530 | 12.15 |
ENSRNOT00000076035
ENSRNOT00000090799 |
Mdn1
|
midasin AAA ATPase 1 |
| chr1_+_61553449 | 10.25 |
ENSRNOT00000068694
|
RGD1565566
|
similar to 60S ribosomal protein L18a |
| chr7_+_143060597 | 9.45 |
ENSRNOT00000087481
|
Krt7
|
keratin 7 |
| chr20_-_6548179 | 8.83 |
ENSRNOT00000093711
|
Lemd2
|
LEM domain containing 2 |
| chr1_+_124983452 | 8.60 |
ENSRNOT00000021262
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr7_-_2961873 | 8.15 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr16_+_61926586 | 7.85 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
| chr5_+_150725654 | 7.78 |
ENSRNOT00000089852
ENSRNOT00000017740 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr20_-_44255064 | 7.76 |
ENSRNOT00000000735
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr12_+_13102019 | 7.71 |
ENSRNOT00000092628
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr7_-_132984110 | 7.53 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr18_+_83777665 | 7.48 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chr8_-_32431987 | 7.30 |
ENSRNOT00000048460
|
RGD1563958
|
similar to 60S ribosomal protein L32 |
| chr1_-_226630900 | 7.23 |
ENSRNOT00000028092
|
Tmem138
|
transmembrane protein 138 |
| chr8_-_62386264 | 7.19 |
ENSRNOT00000026165
|
Cplx3
|
complexin 3 |
| chrX_+_156429585 | 6.91 |
ENSRNOT00000083203
ENSRNOT00000077322 |
Dnase1l1
|
deoxyribonuclease 1-like 1 |
| chr7_+_2458833 | 6.89 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr1_-_170579616 | 6.69 |
ENSRNOT00000076100
|
Rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr18_+_3705916 | 6.55 |
ENSRNOT00000092846
|
Lama3
|
laminin subunit alpha 3 |
| chr6_+_91697109 | 6.42 |
ENSRNOT00000006355
|
Arf6
|
ADP-ribosylation factor 6 |
| chr2_+_230186198 | 6.33 |
ENSRNOT00000013049
ENSRNOT00000092954 |
Casp6
|
caspase 6 |
| chr1_-_94404211 | 6.32 |
ENSRNOT00000019463
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr2_+_235341365 | 6.26 |
ENSRNOT00000078017
ENSRNOT00000090658 |
LOC103689977
|
caspase-6 |
| chr1_-_199336451 | 6.16 |
ENSRNOT00000035672
|
Prss53
|
protease, serine, 53 |
| chr5_-_173659675 | 6.04 |
ENSRNOT00000027547
|
Klhl17
|
kelch-like family member 17 |
| chr11_-_71419223 | 5.90 |
ENSRNOT00000002407
|
Tfrc
|
transferrin receptor |
| chr20_+_13680716 | 5.31 |
ENSRNOT00000029458
|
LOC103694876
|
SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 |
| chr5_+_147069616 | 5.19 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr4_+_140377565 | 5.18 |
ENSRNOT00000082723
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr8_-_133002201 | 5.01 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr12_-_18274515 | 4.99 |
ENSRNOT00000078075
|
LOC100361826
|
sialophorin-like |
| chr3_-_176716146 | 4.78 |
ENSRNOT00000017668
|
Srms
|
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites |
| chr16_-_20278814 | 4.74 |
ENSRNOT00000025385
|
Map1s
|
microtubule-associated protein 1S |
| chr10_-_40994872 | 4.72 |
ENSRNOT00000058640
|
RGD1563668
|
similar to High mobility group protein 1 (HMG-1) |
| chr2_-_116374584 | 4.57 |
ENSRNOT00000058919
|
Lrriq4
|
leucine-rich repeats and IQ motif containing 4 |
| chr9_+_94310921 | 4.53 |
ENSRNOT00000026646
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr14_+_48740190 | 4.44 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_-_65387169 | 4.39 |
ENSRNOT00000051843
|
Orc2
|
origin recognition complex, subunit 2 |
| chr8_+_59111271 | 4.26 |
ENSRNOT00000029413
|
Sh2d7
|
SH2 domain containing 7 |
| chr11_-_71619167 | 4.24 |
ENSRNOT00000002396
|
Tm4sf19
|
transmembrane 4 L six family member 19 |
| chr5_+_135574172 | 4.21 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr6_-_91490366 | 4.05 |
ENSRNOT00000059187
|
Dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr7_-_28932641 | 3.97 |
ENSRNOT00000059487
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr3_-_161131812 | 3.95 |
ENSRNOT00000067008
|
Wfdc11
|
WAP four-disulfide core domain 11 |
| chr8_-_22086095 | 3.90 |
ENSRNOT00000068598
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr12_-_46493203 | 3.84 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr10_+_4719713 | 3.70 |
ENSRNOT00000003412
|
Litaf
|
lipopolysaccharide-induced TNF factor |
| chr12_-_2170504 | 3.44 |
ENSRNOT00000001309
|
Xab2
|
XPA binding protein 2 |
| chr4_+_147455533 | 3.43 |
ENSRNOT00000081957
|
Tsen2
|
tRNA splicing endonuclease subunit 2 |
| chr2_+_187990242 | 3.27 |
ENSRNOT00000092819
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr13_-_55886679 | 3.04 |
ENSRNOT00000066601
|
RGD1566099
|
similar to novel protein |
| chr19_+_53723822 | 3.01 |
ENSRNOT00000023478
|
RGD1304884
|
similar to RIKEN cDNA 6430548M08 |
| chr12_-_22472044 | 2.86 |
ENSRNOT00000073356
|
Ufsp1
|
UFM1-specific peptidase 1 |
| chr1_-_213907144 | 2.81 |
ENSRNOT00000054874
|
Sigirr
|
single Ig and TIR domain containing |
| chr9_-_10659357 | 2.74 |
ENSRNOT00000091450
|
Kdm4b
|
lysine demethylase 4B |
| chr5_+_173340487 | 2.73 |
ENSRNOT00000078306
|
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr20_-_43108198 | 2.68 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr13_+_68949665 | 2.63 |
ENSRNOT00000003320
|
Fam129a
|
family with sequence similarity 129, member A |
| chr2_-_94903226 | 2.60 |
ENSRNOT00000015194
|
AABR07009373.1
|
|
| chr20_+_11863082 | 2.58 |
ENSRNOT00000001640
|
Fam207a
|
family with sequence similarity 207, member A |
| chr1_+_125367280 | 2.27 |
ENSRNOT00000022049
|
Apba2
|
amyloid beta precursor protein binding family A member 2 |
| chr2_+_204886202 | 2.26 |
ENSRNOT00000022200
|
Ngf
|
nerve growth factor |
| chrX_-_20807216 | 2.23 |
ENSRNOT00000075757
|
Fam156b
|
family with sequence similarity 156, member B |
| chr6_-_98606109 | 1.96 |
ENSRNOT00000006860
|
Wdr89
|
WD repeat domain 89 |
| chr4_+_78140483 | 1.96 |
ENSRNOT00000048576
|
Zfp862
|
zinc finger protein 862 |
| chr4_+_168396775 | 1.91 |
ENSRNOT00000008645
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr9_+_94310469 | 1.75 |
ENSRNOT00000065743
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr7_+_125940323 | 1.72 |
ENSRNOT00000083830
|
Fam118a
|
family with sequence similarity 118, member A |
| chr12_-_11294058 | 1.63 |
ENSRNOT00000001319
|
Arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr3_+_80081647 | 1.45 |
ENSRNOT00000057161
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr15_+_42808897 | 1.43 |
ENSRNOT00000023475
|
Chrna2
|
cholinergic receptor nicotinic alpha 2 subunit |
| chr15_+_19690194 | 1.36 |
ENSRNOT00000010612
|
Styx
|
serine/threonine/tyrosine interacting protein |
| chr1_+_220840118 | 1.24 |
ENSRNOT00000027927
|
Fibp
|
FGF1 intracellular binding protein |
| chr5_+_135882967 | 1.18 |
ENSRNOT00000024841
|
Eif2b3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma |
| chr15_-_28835796 | 1.06 |
ENSRNOT00000060439
|
Olr1641
|
olfactory receptor 1641 |
| chr9_+_92859988 | 0.83 |
ENSRNOT00000085332
|
Cab39
|
calcium binding protein 39 |
| chr5_+_18901039 | 0.80 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr10_+_63635219 | 0.80 |
ENSRNOT00000081813
ENSRNOT00000005016 |
Prpf8
|
pre-mRNA processing factor 8 |
| chr2_-_188216482 | 0.73 |
ENSRNOT00000027579
|
Msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr2_-_157066781 | 0.73 |
ENSRNOT00000001187
|
LOC304239
|
similar to RalA binding protein 1 |
| chr3_+_4233111 | 0.72 |
ENSRNOT00000042910
|
Lcn1
|
lipocalin 1 |
| chr11_+_30550141 | 0.67 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr3_+_63536166 | 0.64 |
ENSRNOT00000016132
|
Plekha3
|
pleckstrin homology domain containing A3 |
| chr3_-_22459257 | 0.61 |
ENSRNOT00000091747
|
Dennd1a
|
DENN domain containing 1A |
| chrX_+_157854272 | 0.55 |
ENSRNOT00000089777
ENSRNOT00000003145 |
Fam122b
|
family with sequence similarity 122B |
| chr11_-_28598179 | 0.48 |
ENSRNOT00000070889
|
LOC100912272
|
keratin-associated protein 26-1-like |
| chr13_+_49195325 | 0.46 |
ENSRNOT00000004810
|
Dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr11_-_11214141 | 0.43 |
ENSRNOT00000065748
|
Robo2
|
roundabout guidance receptor 2 |
| chr2_+_20431243 | 0.25 |
ENSRNOT00000021857
|
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr11_-_87403367 | 0.03 |
ENSRNOT00000057751
|
Thap7
|
THAP domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp691
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.6 | 46.2 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 9.4 | 37.8 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 9.1 | 27.4 | GO:2000464 | positive regulation of astrocyte chemotaxis(GO:2000464) |
| 7.4 | 22.2 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 6.9 | 69.2 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 6.5 | 19.5 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 6.1 | 54.5 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 5.0 | 40.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 4.5 | 36.0 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 3.5 | 17.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 3.3 | 20.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 2.5 | 22.8 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 2.3 | 6.9 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 1.9 | 15.0 | GO:0016584 | maintenance of DNA methylation(GO:0010216) nucleosome positioning(GO:0016584) |
| 1.7 | 45.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 1.6 | 24.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.5 | 7.7 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.4 | 14.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.3 | 6.3 | GO:0072733 | acute inflammatory response to non-antigenic stimulus(GO:0002525) response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 1.2 | 19.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 1.1 | 46.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.9 | 2.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.9 | 2.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.9 | 5.2 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.9 | 3.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.8 | 6.7 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.8 | 3.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.8 | 8.8 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.8 | 8.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.7 | 5.9 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.7 | 10.0 | GO:0031498 | chromatin disassembly(GO:0031498) |
| 0.6 | 23.0 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.5 | 19.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.5 | 32.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.5 | 21.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) response to exogenous dsRNA(GO:0043330) |
| 0.4 | 23.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.4 | 6.4 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.3 | 6.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 2.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.3 | 21.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.3 | 31.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.3 | 6.9 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.2 | 5.2 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 3.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 4.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 3.7 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 7.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 15.9 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 18.2 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 7.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 4.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 3.9 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 4.2 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.1 | 6.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 7.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.3 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.7 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 1.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 7.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 4.6 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 2.6 | GO:0001933 | negative regulation of protein phosphorylation(GO:0001933) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 37.8 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.9 | 8.6 | GO:0035838 | growing cell tip(GO:0035838) |
| 1.3 | 6.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.3 | 5.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.1 | 19.1 | GO:0042581 | specific granule(GO:0042581) |
| 1.1 | 6.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 1.0 | 5.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.7 | 19.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.7 | 3.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.7 | 23.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.6 | 7.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 4.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.5 | 1.9 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.5 | 8.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 2.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 6.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.4 | 6.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 6.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 36.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.3 | 91.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 1.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 16.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 5.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 14.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 6.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 7.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 23.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 9.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 15.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 12.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 34.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 62.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 3.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 4.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.6 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 3.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 14.0 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 4.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 9.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 10.9 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 4.0 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 10.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 60.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 34.5 | GO:0044421 | extracellular region part(GO:0044421) |
| 0.0 | 9.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.4 | 46.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 11.5 | 69.2 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 10.8 | 32.4 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 3.9 | 23.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 3.8 | 19.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 3.7 | 22.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 3.0 | 21.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 2.4 | 24.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 2.4 | 18.9 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 2.0 | 6.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.3 | 5.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.0 | 45.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.9 | 2.7 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.9 | 7.7 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.7 | 3.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.7 | 5.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.6 | 2.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.5 | 6.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.4 | 6.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.4 | 4.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 21.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 4.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 2.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 86.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 38.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.3 | 7.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 6.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 39.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 19.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 6.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 7.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 20.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 11.6 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.1 | 8.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 7.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 4.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 28.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 1.4 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 6.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 8.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 3.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 21.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.9 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 9.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 69.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 2.0 | 22.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 1.6 | 24.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 1.3 | 69.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 1.3 | 46.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 1.1 | 65.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 1.0 | 23.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.9 | 20.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.7 | 31.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.7 | 46.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.5 | 19.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 6.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 6.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.3 | 5.2 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 2.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 4.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 19.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 30.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 13.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 27.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 5.4 | 37.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 4.6 | 69.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 2.3 | 41.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.5 | 70.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.2 | 22.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.9 | 54.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 7.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.4 | 4.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 19.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 6.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 5.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 2.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.2 | 26.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 6.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 8.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.8 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 2.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |