Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
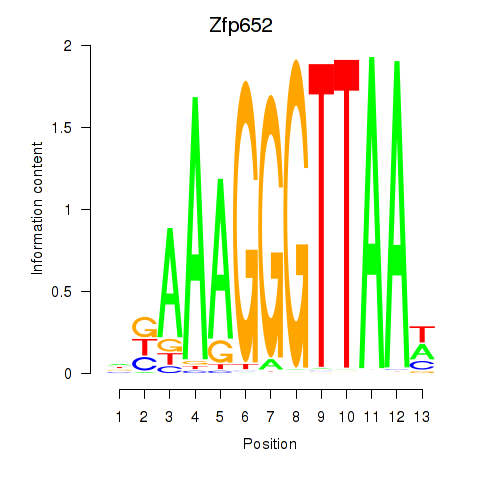
Results for Zfp652
Z-value: 0.94
Transcription factors associated with Zfp652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp652
|
ENSRNOG00000017001 | zinc finger protein 652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp652 | rn6_v1_chr10_+_83556809_83556809 | 0.15 | 5.6e-03 | Click! |
Activity profile of Zfp652 motif
Sorted Z-values of Zfp652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_140483693 | 45.06 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chrX_+_107496072 | 44.51 |
ENSRNOT00000003283
|
Plp1
|
proteolipid protein 1 |
| chr8_+_55178289 | 43.55 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr10_-_51778939 | 29.20 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr8_-_58542844 | 26.67 |
ENSRNOT00000012041
|
Elmod1
|
ELMO domain containing 1 |
| chr6_+_119519714 | 25.52 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_152473868 | 25.29 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr6_-_42473738 | 24.52 |
ENSRNOT00000033327
|
Kcnf1
|
potassium voltage-gated channel modifier subfamily F member 1 |
| chr18_-_61057365 | 22.86 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr2_+_187344056 | 22.48 |
ENSRNOT00000025314
|
Nes
|
nestin |
| chr15_-_28746042 | 22.46 |
ENSRNOT00000017730
|
Sall2
|
spalt-like transcription factor 2 |
| chrX_-_134866210 | 21.44 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr1_-_72727112 | 21.39 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr4_+_114835064 | 21.04 |
ENSRNOT00000031964
|
LOC103692165
|
rhotekin |
| chr16_-_79973735 | 19.94 |
ENSRNOT00000057845
ENSRNOT00000086896 |
Dlgap2
|
DLG associated protein 2 |
| chr3_-_66417741 | 19.35 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr1_+_79989019 | 19.26 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr10_-_109224370 | 19.05 |
ENSRNOT00000005839
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr13_-_88436789 | 18.54 |
ENSRNOT00000003901
|
Ddr2
|
discoidin domain receptor tyrosine kinase 2 |
| chr1_+_157920786 | 18.39 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr10_-_82852660 | 18.09 |
ENSRNOT00000005641
|
Pdk2
|
pyruvate dehydrogenase kinase 2 |
| chr1_-_84254645 | 17.81 |
ENSRNOT00000077651
|
Sptbn4
|
spectrin, beta, non-erythrocytic 4 |
| chr4_+_113968995 | 17.61 |
ENSRNOT00000079511
|
Rtkn
|
rhotekin |
| chr13_-_84331905 | 16.88 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr10_+_53466870 | 16.31 |
ENSRNOT00000057503
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr8_-_117237229 | 15.99 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr17_-_15467320 | 15.82 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr18_+_61261418 | 15.73 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr9_-_113504605 | 15.57 |
ENSRNOT00000067974
|
Rab31
|
RAB31, member RAS oncogene family |
| chr10_+_31146107 | 15.39 |
ENSRNOT00000008184
|
Adam19
|
ADAM metallopeptidase domain 19 |
| chr7_+_78558701 | 15.16 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr13_+_60435946 | 15.05 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr10_-_83332851 | 14.91 |
ENSRNOT00000007133
|
Nxph3
|
neurexophilin 3 |
| chr2_+_50099576 | 14.79 |
ENSRNOT00000089218
|
Hcn1
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 |
| chr4_+_140837745 | 14.66 |
ENSRNOT00000080663
|
Arl8b
|
ADP-ribosylation factor like GTPase 8B |
| chr17_-_14717420 | 14.39 |
ENSRNOT00000071131
|
LOC100910978
|
extracellular matrix protein 2-like |
| chr11_-_87017115 | 14.21 |
ENSRNOT00000051037
|
Rtn4r
|
reticulon 4 receptor |
| chr15_-_60766579 | 13.48 |
ENSRNOT00000079978
|
Akap11
|
A-kinase anchoring protein 11 |
| chr1_+_150797084 | 12.96 |
ENSRNOT00000018990
|
Nox4
|
NADPH oxidase 4 |
| chr4_+_7355574 | 12.68 |
ENSRNOT00000013800
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr17_-_45154355 | 11.53 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr10_+_56627411 | 11.33 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr8_+_63600663 | 10.99 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr4_-_84768249 | 10.94 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
| chr17_-_15555919 | 10.61 |
ENSRNOT00000043852
|
Ecm2
|
extracellular matrix protein 2 |
| chr6_-_51018050 | 10.59 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr13_+_104284660 | 10.41 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr3_+_45277348 | 10.24 |
ENSRNOT00000059246
|
Pkp4
|
plakophilin 4 |
| chrX_+_82143789 | 9.98 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr11_+_74834050 | 9.57 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr3_-_162581610 | 9.27 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr8_+_22189600 | 9.11 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chr8_-_63350269 | 8.60 |
ENSRNOT00000042681
|
Cd276
|
Cd276 molecule |
| chr5_+_79055521 | 8.45 |
ENSRNOT00000010333
|
Col27a1
|
collagen type XXVII alpha 1 chain |
| chr1_+_199664173 | 7.46 |
ENSRNOT00000054980
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr10_+_18912865 | 7.28 |
ENSRNOT00000007393
|
Kcnmb1
|
potassium calcium-activated channel subfamily M regulatory beta subunit 1 |
| chr10_-_86645529 | 6.75 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr3_-_175601127 | 6.72 |
ENSRNOT00000081226
|
Lama5
|
laminin subunit alpha 5 |
| chr11_+_57404196 | 6.67 |
ENSRNOT00000042754
ENSRNOT00000034097 |
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr1_-_248898607 | 6.16 |
ENSRNOT00000074393
|
NEWGENE_1307313
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr13_-_103080920 | 6.12 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr13_+_71107465 | 5.45 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr10_-_81942188 | 5.43 |
ENSRNOT00000003784
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr12_-_31249118 | 5.42 |
ENSRNOT00000058586
ENSRNOT00000073105 |
Adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr1_-_239057732 | 4.41 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr9_+_93326283 | 4.01 |
ENSRNOT00000024578
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr6_-_102196138 | 3.81 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr6_+_99625306 | 3.67 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr8_-_71052529 | 3.63 |
ENSRNOT00000033512
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr13_-_85443727 | 3.25 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr7_-_123088819 | 2.78 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr1_-_43638161 | 2.74 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_117259141 | 2.43 |
ENSRNOT00000040762
|
Plec
|
plectin |
| chr14_-_44845218 | 2.28 |
ENSRNOT00000004003
|
Klhl5
|
kelch-like family member 5 |
| chr6_-_108596446 | 2.25 |
ENSRNOT00000031331
ENSRNOT00000077458 ENSRNOT00000082792 |
Ltbp2
|
latent transforming growth factor beta binding protein 2 |
| chr12_+_22835019 | 2.18 |
ENSRNOT00000059530
|
Col26a1
|
collagen type XXVI alpha 1 chain |
| chr3_-_113423149 | 1.86 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr2_-_189856090 | 1.77 |
ENSRNOT00000020307
|
Npr1
|
natriuretic peptide receptor 1 |
| chr8_+_117648745 | 1.66 |
ENSRNOT00000027762
|
Slc26a6
|
solute carrier family 26 member 6 |
| chr1_+_229461127 | 1.53 |
ENSRNOT00000035813
|
Olr332
|
olfactory receptor 332 |
| chr1_-_84040433 | 0.86 |
ENSRNOT00000018696
|
Itpkc
|
inositol-trisphosphate 3-kinase C |
| chr6_+_76383446 | 0.39 |
ENSRNOT00000010188
|
Insm2
|
INSM transcriptional repressor 2 |
| chr10_+_12929471 | 0.17 |
ENSRNOT00000036563
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr4_+_157554794 | 0.03 |
ENSRNOT00000024116
|
Ing4
|
inhibitor of growth family, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 5.6 | 22.5 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 5.4 | 43.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 5.4 | 21.4 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 4.5 | 18.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 4.3 | 25.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 4.2 | 12.7 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 3.9 | 19.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 3.9 | 19.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 3.7 | 14.8 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.9 | 8.6 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 2.8 | 31.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 2.6 | 10.4 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 2.5 | 37.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 2.4 | 14.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 2.3 | 9.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 2.2 | 11.0 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 2.1 | 8.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 2.0 | 17.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.9 | 18.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 1.8 | 7.3 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.7 | 6.7 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.2 | 15.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.1 | 19.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 1.1 | 15.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.1 | 7.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.0 | 38.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.9 | 4.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.8 | 15.6 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.7 | 13.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.7 | 22.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.7 | 10.0 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.6 | 1.7 | GO:0046724 | oxalic acid secretion(GO:0046724) |
| 0.5 | 16.3 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.5 | 5.4 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.5 | 5.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 1.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.3 | 21.4 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.3 | 15.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 17.6 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.2 | 10.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 6.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 10.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 14.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 1.9 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 14.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 2.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 6.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 19.6 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 2.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 3.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 14.9 | GO:0021915 | neural tube development(GO:0021915) |
| 0.1 | 6.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 6.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 16.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 11.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 2.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 2.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.8 | GO:0098855 | HCN channel complex(GO:0098855) |
| 6.0 | 18.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 4.8 | 43.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.0 | 45.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 2.2 | 17.8 | GO:0043203 | axon hillock(GO:0043203) |
| 1.7 | 6.7 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 1.3 | 13.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.3 | 11.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.2 | 19.9 | GO:0005883 | neurofilament(GO:0005883) |
| 1.0 | 22.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.8 | 14.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.7 | 24.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.7 | 8.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.7 | 19.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.6 | 15.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 6.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.4 | 19.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 44.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 15.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 18.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 4.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 5.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 2.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 44.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 60.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.2 | 22.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 9.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 21.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 26.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 19.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 5.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 22.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 6.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 4.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 12.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 4.8 | 38.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 4.5 | 18.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 3.7 | 44.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.1 | 31.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 3.0 | 15.0 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 2.6 | 18.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.6 | 25.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 2.1 | 25.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 2.0 | 43.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.9 | 22.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.8 | 12.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.8 | 14.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.6 | 21.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 1.5 | 7.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.4 | 13.0 | GO:0016175 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.0 | 29.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 1.0 | 10.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 10.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 10.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 13.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.6 | 1.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.6 | 14.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.6 | 1.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.5 | 21.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.5 | 20.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.5 | 9.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.5 | 19.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 16.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.4 | 9.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 9.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 16.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 1.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 19.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.3 | 7.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 15.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 19.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 6.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 4.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.2 | 1.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 5.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 8.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.0 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 19.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 15.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 15.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 26.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 22.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 5.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 15.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 6.7 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 15.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 11.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 21.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 2.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 29.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 13.0 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 10.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 10.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 18.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.1 | 15.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 19.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.7 | 37.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.6 | 17.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 33.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.4 | 4.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 10.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 4.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 8.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 6.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |