Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
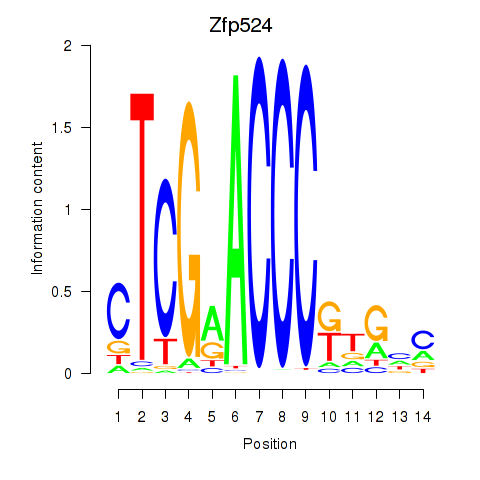
Results for Zfp524
Z-value: 1.19
Transcription factors associated with Zfp524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp524
|
ENSRNOG00000016565 | zinc finger protein 524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp524 | rn6_v1_chr1_-_72377434_72377434 | 0.45 | 3.6e-17 | Click! |
Activity profile of Zfp524 motif
Sorted Z-values of Zfp524 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_89386023 | 31.94 |
ENSRNOT00000086223
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr13_+_89385859 | 29.46 |
ENSRNOT00000047434
|
Fcgr3a
|
Fc fragment of IgG receptor IIIa |
| chr3_-_80012750 | 26.58 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr19_+_24456976 | 26.49 |
ENSRNOT00000004900
|
Ucp1
|
uncoupling protein 1 |
| chr4_+_99239115 | 25.55 |
ENSRNOT00000009515
|
Cd8a
|
CD8a molecule |
| chr1_-_73619356 | 24.78 |
ENSRNOT00000074352
|
Lilrb3
|
leukocyte immunoglobulin like receptor B3 |
| chr17_+_24416651 | 24.14 |
ENSRNOT00000024458
|
Cd83
|
CD83 molecule |
| chr13_+_83996080 | 22.71 |
ENSRNOT00000004403
ENSRNOT00000070958 |
Cd247
|
Cd247 molecule |
| chr13_-_50499060 | 21.83 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr9_-_9985358 | 20.51 |
ENSRNOT00000080856
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr20_+_3990820 | 20.20 |
ENSRNOT00000000528
|
Psmb8
|
proteasome subunit beta 8 |
| chr8_-_62424303 | 19.54 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr10_-_70341837 | 19.14 |
ENSRNOT00000077261
|
Slfn13
|
schlafen family member 13 |
| chr1_-_85317968 | 19.06 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr8_+_22035256 | 18.84 |
ENSRNOT00000028066
|
Icam1
|
intercellular adhesion molecule 1 |
| chr4_-_157252565 | 18.79 |
ENSRNOT00000079947
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_-_70499200 | 18.45 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr5_+_137255924 | 18.09 |
ENSRNOT00000088022
ENSRNOT00000092755 |
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr3_-_14112851 | 17.84 |
ENSRNOT00000092736
|
C5
|
complement C5 |
| chr10_-_88442796 | 17.56 |
ENSRNOT00000077579
ENSRNOT00000023447 ENSRNOT00000043172 |
Acly
|
ATP citrate lyase |
| chr1_-_101095594 | 17.36 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr19_-_838099 | 16.61 |
ENSRNOT00000014234
|
Cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr10_+_63803309 | 16.50 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chr4_-_163402561 | 16.31 |
ENSRNOT00000091890
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr2_-_23256158 | 16.30 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr13_-_47979797 | 16.07 |
ENSRNOT00000080035
|
Rassf5
|
Ras association domain family member 5 |
| chr1_+_145770135 | 15.86 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr9_-_10471009 | 15.73 |
ENSRNOT00000072868
|
Safb
|
scaffold attachment factor B |
| chr14_-_82287706 | 15.51 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr3_+_18706988 | 15.35 |
ENSRNOT00000074650
|
AABR07051652.1
|
|
| chr17_+_9639330 | 15.01 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr11_+_87204175 | 14.74 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr20_-_5212624 | 14.55 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr10_+_86340940 | 14.53 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr6_+_49968473 | 14.42 |
ENSRNOT00000007466
|
Fam110c
|
family with sequence similarity 110, member C |
| chr10_+_15241590 | 13.96 |
ENSRNOT00000037372
ENSRNOT00000037381 |
Mettl26
|
methyltransferase like 26 |
| chr4_+_152995865 | 13.82 |
ENSRNOT00000014885
|
Il17ra
|
interleukin 17 receptor A |
| chr19_-_19376789 | 13.82 |
ENSRNOT00000065103
|
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr8_+_49282460 | 13.72 |
ENSRNOT00000021488
|
Cd3d
|
CD3d molecule |
| chr10_+_109707962 | 13.68 |
ENSRNOT00000054962
ENSRNOT00000088898 |
Gcgr
|
glucagon receptor |
| chr6_-_11298216 | 13.66 |
ENSRNOT00000021135
|
Epcam
|
epithelial cell adhesion molecule |
| chr13_+_52976507 | 13.49 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr8_-_22336794 | 13.43 |
ENSRNOT00000066340
|
Ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr7_-_73270308 | 13.42 |
ENSRNOT00000007430
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr3_-_113405829 | 13.14 |
ENSRNOT00000036823
|
Ell3
|
elongation factor for RNA polymerase II 3 |
| chr8_+_67295727 | 12.93 |
ENSRNOT00000020443
|
Anp32a
|
acidic nuclear phosphoprotein 32 family member A |
| chr11_+_47243342 | 12.51 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr10_-_55965216 | 12.46 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr7_-_3170146 | 12.38 |
ENSRNOT00000036373
|
Dgka
|
diacylglycerol kinase, alpha |
| chr1_+_191829555 | 12.23 |
ENSRNOT00000067138
|
Scnn1b
|
sodium channel epithelial 1 beta subunit |
| chr4_+_171748273 | 12.07 |
ENSRNOT00000009998
|
Dera
|
deoxyribose-phosphate aldolase |
| chr10_+_3218466 | 12.03 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr20_+_3744395 | 11.88 |
ENSRNOT00000086065
|
Tcf19
|
transcription factor 19 |
| chr2_-_47096961 | 11.76 |
ENSRNOT00000077401
|
Itga2
|
integrin alpha 2 |
| chr8_-_61919874 | 11.76 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr8_-_128026841 | 11.72 |
ENSRNOT00000018341
|
Myd88
|
myeloid differentiation primary response 88 |
| chr7_+_97984862 | 11.58 |
ENSRNOT00000008508
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr3_-_55822551 | 11.57 |
ENSRNOT00000082624
|
Lrp2
|
LDL receptor related protein 2 |
| chr3_+_66193059 | 11.56 |
ENSRNOT00000006880
|
Itga4
|
integrin subunit alpha 4 |
| chr3_-_10375826 | 11.51 |
ENSRNOT00000093365
|
Ass1
|
argininosuccinate synthase 1 |
| chr6_-_51356383 | 11.46 |
ENSRNOT00000012415
|
Prkar2b
|
protein kinase cAMP-dependent type 2 regulatory subunit beta |
| chr19_+_54314865 | 11.46 |
ENSRNOT00000024069
|
Irf8
|
interferon regulatory factor 8 |
| chr11_+_47260641 | 11.45 |
ENSRNOT00000078856
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr10_-_91448477 | 11.44 |
ENSRNOT00000038836
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr13_+_89774764 | 11.43 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr20_-_45259928 | 11.38 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr4_-_119714302 | 11.36 |
ENSRNOT00000013299
|
Rab43
|
RAB43, member RAS oncogene family |
| chr6_-_45669148 | 11.30 |
ENSRNOT00000010092
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr8_+_11888591 | 11.17 |
ENSRNOT00000083967
|
Ccdc82
|
coiled-coil domain containing 82 |
| chr18_-_28425944 | 11.13 |
ENSRNOT00000084372
|
Slc23a1
|
solute carrier family 23 member 1 |
| chrX_+_123205869 | 11.00 |
ENSRNOT00000017101
ENSRNOT00000079231 |
Pgrmc1
|
progesterone receptor membrane component 1 |
| chr7_+_2605719 | 11.00 |
ENSRNOT00000018737
|
Gls2
|
glutaminase 2 |
| chr1_-_88881460 | 11.00 |
ENSRNOT00000028287
|
Hcst
|
hematopoietic cell signal transducer |
| chr4_-_69268336 | 10.98 |
ENSRNOT00000018042
|
Prss3b
|
protease, serine, 3B |
| chr1_+_217018916 | 10.88 |
ENSRNOT00000028195
ENSRNOT00000078979 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr1_+_16910069 | 10.81 |
ENSRNOT00000020015
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr9_+_20765296 | 10.71 |
ENSRNOT00000016291
|
Cd2ap
|
CD2-associated protein |
| chr11_-_80981415 | 10.51 |
ENSRNOT00000002499
ENSRNOT00000002496 |
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr10_-_70744315 | 10.49 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr5_+_154311998 | 10.45 |
ENSRNOT00000084085
|
Gale
|
UDP-galactose-4-epimerase |
| chr1_-_72941869 | 10.35 |
ENSRNOT00000032306
|
Eps8l1
|
EPS8-like 1 |
| chr4_+_109467272 | 10.35 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chrX_-_104791210 | 10.33 |
ENSRNOT00000004921
|
Sytl4
|
synaptotagmin-like 4 |
| chr7_+_41475163 | 10.25 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr1_+_226687258 | 10.22 |
ENSRNOT00000079679
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr5_+_172648950 | 10.19 |
ENSRNOT00000055361
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr16_+_1749191 | 10.12 |
ENSRNOT00000014004
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr5_-_152122542 | 10.10 |
ENSRNOT00000068634
ENSRNOT00000077248 |
Rps6ka1
|
ribosomal protein S6 kinase A1 |
| chr15_+_40937708 | 10.09 |
ENSRNOT00000018407
|
Spata13
|
spermatogenesis associated 13 |
| chr14_-_33164141 | 10.04 |
ENSRNOT00000002844
|
Rest
|
RE1-silencing transcription factor |
| chr19_+_24800072 | 10.03 |
ENSRNOT00000005470
|
Ptger1
|
prostaglandin E receptor 1 |
| chr15_+_28074953 | 10.03 |
ENSRNOT00000086427
|
LOC103690354
|
ribonuclease pancreatic beta-type |
| chr13_+_49074644 | 10.01 |
ENSRNOT00000000041
|
Klhdc8a
|
kelch domain containing 8A |
| chr20_-_30749940 | 9.98 |
ENSRNOT00000065127
|
Sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr2_-_230273709 | 9.94 |
ENSRNOT00000012587
|
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr4_-_153465203 | 9.90 |
ENSRNOT00000016776
|
Bid
|
BH3 interacting domain death agonist |
| chr2_-_200513564 | 9.90 |
ENSRNOT00000056173
|
Phgdh
|
phosphoglycerate dehydrogenase |
| chr6_+_132762280 | 9.85 |
ENSRNOT00000004070
|
Slc25a47
|
solute carrier family 25, member 47 |
| chr7_-_107616038 | 9.83 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr20_+_3995544 | 9.83 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr10_-_105116916 | 9.70 |
ENSRNOT00000012373
|
Evpl
|
envoplakin |
| chr1_-_198486157 | 9.67 |
ENSRNOT00000022758
|
Zg16
|
zymogen granule protein 16 |
| chr19_-_15540773 | 9.60 |
ENSRNOT00000022359
|
Lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr16_+_19800463 | 9.49 |
ENSRNOT00000023065
|
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr19_-_10513349 | 9.45 |
ENSRNOT00000061270
|
Adgrg5
|
adhesion G protein-coupled receptor G5 |
| chr12_-_16126953 | 9.42 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_78833157 | 9.35 |
ENSRNOT00000022461
|
Ptgir
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr7_-_119768082 | 9.30 |
ENSRNOT00000009612
|
Sstr3
|
somatostatin receptor 3 |
| chr10_+_75149814 | 9.28 |
ENSRNOT00000011500
|
Mks1
|
Meckel syndrome, type 1 |
| chr12_-_41448668 | 9.20 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr1_-_191007503 | 9.15 |
ENSRNOT00000023262
|
Igsf6
|
immunoglobulin superfamily, member 6 |
| chr7_+_116355698 | 9.12 |
ENSRNOT00000076693
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr10_+_76343847 | 9.10 |
ENSRNOT00000055674
|
Trim25
|
tripartite motif-containing 25 |
| chr10_+_4578469 | 9.07 |
ENSRNOT00000003332
|
Txndc11
|
thioredoxin domain containing 11 |
| chr6_+_29012207 | 9.06 |
ENSRNOT00000079077
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr11_+_73936750 | 8.97 |
ENSRNOT00000002350
|
Atp13a3
|
ATPase 13A3 |
| chr3_-_81282157 | 8.97 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr10_-_70342411 | 8.95 |
ENSRNOT00000076269
ENSRNOT00000076477 |
Slfn13
|
schlafen family member 13 |
| chr15_+_24141651 | 8.83 |
ENSRNOT00000082304
|
Lgals3
|
galectin 3 |
| chr5_+_156215417 | 8.74 |
ENSRNOT00000067616
ENSRNOT00000077742 |
Ece1
|
endothelin converting enzyme 1 |
| chr10_-_72545141 | 8.72 |
ENSRNOT00000033684
|
Appbp2
|
amyloid beta precursor protein binding protein 2 |
| chr3_+_161298962 | 8.61 |
ENSRNOT00000066028
|
Ctsa
|
cathepsin A |
| chr12_+_23473270 | 8.61 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr7_+_116355911 | 8.47 |
ENSRNOT00000076182
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr10_+_84182118 | 8.27 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr5_-_139853247 | 8.24 |
ENSRNOT00000056644
|
Exo5
|
exonuclease 5 |
| chr1_+_137014272 | 8.22 |
ENSRNOT00000014802
|
Akap13
|
A-kinase anchoring protein 13 |
| chr10_-_94352880 | 8.21 |
ENSRNOT00000035973
|
Limd2
|
LIM domain containing 2 |
| chr12_-_41590244 | 8.19 |
ENSRNOT00000038736
|
Slc8b1
|
solute carrier family 8 member B1 |
| chr1_-_89474252 | 8.18 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr5_-_24255606 | 8.15 |
ENSRNOT00000037483
|
Plekhf2
|
pleckstrin homology and FYVE domain containing 2 |
| chr1_-_89042176 | 8.10 |
ENSRNOT00000080842
|
Kmt2b
|
lysine methyltransferase 2B |
| chr20_+_46199981 | 8.04 |
ENSRNOT00000000337
|
Mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr10_+_87774552 | 8.02 |
ENSRNOT00000044342
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr7_+_70612103 | 7.96 |
ENSRNOT00000057833
|
Arhgap9
|
Rho GTPase activating protein 9 |
| chr2_+_212247451 | 7.95 |
ENSRNOT00000027813
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr4_+_118207862 | 7.95 |
ENSRNOT00000085787
ENSRNOT00000023103 |
Tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr1_+_235166718 | 7.87 |
ENSRNOT00000020201
|
Gna14
|
G protein subunit alpha 14 |
| chr13_+_83073866 | 7.85 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr16_-_20383337 | 7.79 |
ENSRNOT00000025977
|
Il12rb1
|
interleukin 12 receptor subunit beta 1 |
| chr12_-_46718355 | 7.76 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr20_-_2212171 | 7.72 |
ENSRNOT00000085912
|
Trim26
|
tripartite motif-containing 26 |
| chr1_+_277355619 | 7.72 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr7_+_11301957 | 7.63 |
ENSRNOT00000027847
|
Tjp3
|
tight junction protein 3 |
| chr12_-_1512130 | 7.62 |
ENSRNOT00000086278
|
AABR07034940.2
|
|
| chr15_-_42517553 | 7.61 |
ENSRNOT00000081500
ENSRNOT00000021320 |
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr2_+_225005019 | 7.57 |
ENSRNOT00000015579
|
Cnn3
|
calponin 3 |
| chr7_+_119820537 | 7.55 |
ENSRNOT00000077256
ENSRNOT00000056221 |
Cyth4
|
cytohesin 4 |
| chr1_+_31967978 | 7.52 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr4_-_113886994 | 7.48 |
ENSRNOT00000037333
|
Htra2
|
HtrA serine peptidase 2 |
| chr16_+_20039969 | 7.44 |
ENSRNOT00000024866
|
Colgalt1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr7_-_24004774 | 7.43 |
ENSRNOT00000007549
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr1_-_221041401 | 7.40 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr1_-_80056574 | 7.35 |
ENSRNOT00000021200
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr8_+_49354115 | 7.34 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr19_+_41710102 | 7.33 |
ENSRNOT00000021866
ENSRNOT00000079730 |
Marveld3
|
MARVEL domain containing 3 |
| chr7_+_2458264 | 7.26 |
ENSRNOT00000003550
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr1_+_157701089 | 7.23 |
ENSRNOT00000014271
|
Prcp
|
prolylcarboxypeptidase |
| chr19_+_15094309 | 7.22 |
ENSRNOT00000083500
|
Ces1f
|
carboxylesterase 1F |
| chr12_-_38274036 | 7.17 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr6_+_29012007 | 7.14 |
ENSRNOT00000085612
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr3_+_72385666 | 7.12 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr8_-_7426611 | 7.05 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr6_+_107517668 | 6.97 |
ENSRNOT00000013753
|
Acot4
|
acyl-CoA thioesterase 4 |
| chr5_-_76632462 | 6.95 |
ENSRNOT00000085109
|
Susd1
|
sushi domain containing 1 |
| chr10_-_13898855 | 6.91 |
ENSRNOT00000004249
|
Rab26
|
RAB26, member RAS oncogene family |
| chr1_-_80185882 | 6.89 |
ENSRNOT00000022214
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr9_-_14599594 | 6.88 |
ENSRNOT00000018138
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr20_-_41209728 | 6.87 |
ENSRNOT00000000657
|
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr18_-_69784099 | 6.87 |
ENSRNOT00000049759
|
Me2
|
malic enzyme 2 |
| chr10_-_57064600 | 6.87 |
ENSRNOT00000032926
|
Cxcl16
|
C-X-C motif chemokine ligand 16 |
| chr1_-_261986759 | 6.86 |
ENSRNOT00000021257
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr1_-_98570949 | 6.83 |
ENSRNOT00000033648
|
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr6_-_91518996 | 6.82 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr1_+_91857057 | 6.79 |
ENSRNOT00000077993
ENSRNOT00000081465 |
Ankrd27
|
ankyrin repeat domain 27 |
| chr4_+_34282625 | 6.76 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr4_-_34282351 | 6.74 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chr19_+_58746313 | 6.72 |
ENSRNOT00000026986
|
Map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr20_-_5192000 | 6.69 |
ENSRNOT00000079677
|
Tnf
|
tumor necrosis factor |
| chr4_-_70747226 | 6.61 |
ENSRNOT00000044960
|
LOC102554637
|
anionic trypsin-2-like |
| chr4_-_77489535 | 6.60 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr20_+_3351303 | 6.60 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr1_+_279798187 | 6.57 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr3_+_11554457 | 6.56 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr4_-_163214678 | 6.55 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr5_-_152358643 | 6.51 |
ENSRNOT00000021734
|
Sh3bgrl3
|
SH3 domain binding glutamate-rich protein like 3 |
| chr3_+_111422267 | 6.51 |
ENSRNOT00000052312
|
Nusap1
|
nucleolar and spindle associated protein 1 |
| chr1_+_260093641 | 6.48 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr2_-_27500654 | 6.45 |
ENSRNOT00000022055
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr2_-_182038178 | 6.43 |
ENSRNOT00000040708
|
Fgb
|
fibrinogen beta chain |
| chr5_-_164348713 | 6.37 |
ENSRNOT00000087044
|
LOC500584
|
similar to casein kinase 1, gamma 3 isoform 2 |
| chr10_+_73333119 | 6.35 |
ENSRNOT00000004736
|
Tbx4
|
T-box 4 |
| chr10_-_62287189 | 6.30 |
ENSRNOT00000004365
|
Wdr81
|
WD repeat domain 81 |
| chr1_+_88885937 | 6.27 |
ENSRNOT00000029719
|
Nfkbid
|
NFKB inhibitor delta |
| chr10_-_109840047 | 6.26 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr9_-_9985630 | 6.25 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr1_-_98521551 | 6.24 |
ENSRNOT00000081922
|
Siglec10
|
sialic acid binding Ig-like lectin 10 |
| chr11_-_77593171 | 6.22 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr10_+_88987558 | 6.21 |
ENSRNOT00000026878
|
Hsd17b1
|
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr3_-_3700200 | 6.17 |
ENSRNOT00000036231
|
AC129824.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp524
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.5 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 5.5 | 22.1 | GO:0050904 | diapedesis(GO:0050904) |
| 5.5 | 16.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 5.4 | 16.3 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 5.4 | 16.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 5.3 | 16.0 | GO:0002254 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 5.3 | 26.6 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 5.3 | 78.8 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 5.1 | 25.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 4.9 | 9.8 | GO:0002585 | positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) |
| 4.7 | 18.8 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 4.6 | 13.8 | GO:0002777 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 4.1 | 12.3 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 3.8 | 11.5 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 3.8 | 19.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 3.8 | 11.3 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 3.8 | 11.3 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 3.7 | 11.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 3.7 | 14.7 | GO:0015746 | citrate transport(GO:0015746) |
| 3.6 | 25.5 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 3.6 | 10.9 | GO:0016129 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 3.6 | 14.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 3.5 | 10.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 3.4 | 24.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 3.4 | 13.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 3.3 | 19.8 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 3.3 | 19.7 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 3.2 | 9.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 3.1 | 15.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 3.0 | 12.1 | GO:1903576 | response to L-arginine(GO:1903576) |
| 3.0 | 17.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.9 | 8.8 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 2.9 | 8.7 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 2.7 | 8.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 2.7 | 10.7 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 2.6 | 7.8 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 2.6 | 18.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 2.5 | 10.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 2.5 | 27.8 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 2.5 | 10.0 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 2.5 | 12.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 2.5 | 5.0 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 2.5 | 9.8 | GO:0015879 | carnitine transport(GO:0015879) |
| 2.5 | 7.4 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 2.4 | 12.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 2.4 | 7.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.4 | 7.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 2.3 | 7.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 2.3 | 11.7 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 2.3 | 11.4 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 2.3 | 6.8 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 2.2 | 6.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 2.1 | 4.2 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 2.1 | 16.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 2.1 | 6.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 2.1 | 8.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 2.1 | 8.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 2.0 | 6.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 2.0 | 6.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 2.0 | 6.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 1.9 | 7.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.9 | 9.5 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 1.9 | 5.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.8 | 27.5 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 1.8 | 14.5 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 1.8 | 5.4 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 1.8 | 7.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 1.7 | 10.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 1.7 | 5.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 1.7 | 28.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 1.7 | 11.8 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 1.7 | 5.0 | GO:0006407 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 1.6 | 13.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 1.6 | 14.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.6 | 9.3 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 1.5 | 1.5 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 1.5 | 13.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.5 | 4.4 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 1.5 | 21.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 1.4 | 5.7 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 1.4 | 4.3 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 1.4 | 8.6 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.4 | 5.7 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 1.4 | 13.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.4 | 4.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 1.4 | 13.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 1.4 | 4.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.4 | 4.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 1.4 | 6.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 1.4 | 2.7 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 1.3 | 5.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.3 | 6.6 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 1.3 | 10.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 1.3 | 6.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.3 | 21.9 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 1.3 | 3.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.3 | 3.8 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 1.3 | 6.3 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 1.2 | 7.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 1.2 | 3.7 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.2 | 9.9 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 1.2 | 6.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 1.2 | 8.6 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 1.2 | 4.8 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 1.2 | 3.6 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 1.2 | 5.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.2 | 3.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 1.2 | 4.6 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 1.1 | 4.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.1 | 7.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 1.1 | 5.5 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 1.1 | 7.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.1 | 7.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.1 | 15.0 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 1.1 | 5.3 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) |
| 1.1 | 7.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 1.1 | 15.9 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 1.1 | 5.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 1.0 | 2.1 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) |
| 1.0 | 10.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 1.0 | 4.0 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 1.0 | 4.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 1.0 | 12.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 1.0 | 4.8 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.9 | 6.6 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.9 | 1.9 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.9 | 3.6 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.9 | 2.7 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.9 | 8.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.9 | 1.8 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.9 | 24.8 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.9 | 17.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.9 | 2.6 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.9 | 6.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.9 | 6.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.9 | 6.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.8 | 4.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.8 | 16.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.8 | 2.5 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 0.8 | 12.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.8 | 10.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.8 | 2.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.8 | 4.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.8 | 6.9 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.8 | 2.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.7 | 3.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.7 | 2.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.7 | 1.5 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.7 | 11.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.7 | 10.1 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.7 | 3.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.7 | 6.3 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.7 | 4.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.7 | 6.2 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.7 | 4.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.7 | 12.9 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.7 | 10.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.7 | 3.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.7 | 2.6 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.7 | 5.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.7 | 4.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.6 | 1.3 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.6 | 10.0 | GO:0030149 | sphingolipid catabolic process(GO:0030149) |
| 0.6 | 3.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.6 | 3.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.6 | 3.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.6 | 3.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.6 | 1.2 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.6 | 7.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.6 | 10.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) |
| 0.6 | 11.4 | GO:0019068 | virion assembly(GO:0019068) |
| 0.6 | 3.0 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.6 | 9.4 | GO:0043950 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.6 | 1.7 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.6 | 2.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.6 | 2.9 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.6 | 5.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.6 | 3.4 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.6 | 3.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.6 | 3.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.6 | 1.7 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.5 | 3.3 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.5 | 8.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 3.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 | 5.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.5 | 1.0 | GO:1990268 | response to gold nanoparticle(GO:1990268) |
| 0.5 | 2.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.5 | 6.2 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.5 | 1.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.5 | 9.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.5 | 7.6 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 4.4 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.5 | 3.9 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.5 | 4.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.5 | 6.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 8.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.5 | 4.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 1.9 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 4.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 2.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.5 | 1.8 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.5 | 5.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.5 | 10.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 1.8 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 3.9 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.4 | 5.7 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.4 | 5.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 | 11.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.4 | 5.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.4 | 4.6 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.4 | 0.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 | 1.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.4 | 3.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.4 | 4.1 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.4 | 5.7 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.4 | 3.6 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.4 | 1.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 12.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.4 | 4.7 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.4 | 1.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.4 | 2.3 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.4 | 3.5 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.4 | 27.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.4 | 12.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.4 | 1.5 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.4 | 5.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.4 | 1.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 1.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.4 | 4.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.4 | 1.1 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.4 | 20.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.3 | 1.0 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 1.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 | 2.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.3 | 1.9 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 | 3.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 8.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.3 | 10.0 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.3 | 9.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 1.5 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.3 | 4.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 3.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.3 | 3.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 7.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.3 | 4.1 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.3 | 24.0 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.3 | 4.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.3 | 5.4 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.3 | 1.4 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.3 | 1.4 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 2.8 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.3 | 2.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 18.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.3 | 5.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.3 | 7.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.3 | 0.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 2.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.3 | 3.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 0.8 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.3 | 1.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 5.8 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.3 | 0.5 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.3 | 1.6 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.2 | 5.0 | GO:0007567 | parturition(GO:0007567) |
| 0.2 | 1.9 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 6.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.5 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 0.9 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.2 | 3.0 | GO:0015816 | glycine transport(GO:0015816) |
| 0.2 | 1.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.9 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 0.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 5.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 2.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 10.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 3.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 2.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 3.3 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 0.8 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 4.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 | 0.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 0.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 | 5.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.2 | 0.8 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 1.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.1 | GO:0044351 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) macropinocytosis(GO:0044351) |
| 0.2 | 1.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 1.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 2.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 2.5 | GO:2000641 | regulation of early endosome to late endosome transport(GO:2000641) |
| 0.2 | 1.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.2 | 6.8 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.2 | 4.7 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.2 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.5 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.2 | 3.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 4.6 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.2 | 8.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 3.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 4.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 8.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.2 | 0.3 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) |
| 0.1 | 1.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 6.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 5.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.8 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 1.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 6.9 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 1.9 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 3.6 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 9.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 11.0 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 2.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 11.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 5.1 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 5.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 2.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 3.5 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.5 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 3.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.6 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.1 | 2.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.9 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 3.8 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.1 | 3.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.2 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.4 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 | 2.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 3.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 3.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.6 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 3.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.3 | GO:0006311 | meiotic gene conversion(GO:0006311) male meiosis chromosome segregation(GO:0007060) gene conversion(GO:0035822) |
| 0.1 | 2.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 7.6 | GO:0048705 | skeletal system morphogenesis(GO:0048705) |
| 0.0 | 0.7 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 1.3 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.9 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 5.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 4.1 | GO:1900180 | regulation of protein localization to nucleus(GO:1900180) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 1.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.1 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 2.1 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 3.0 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.5 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 1.3 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 55.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 3.9 | 11.8 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 3.5 | 24.4 | GO:0042825 | TAP complex(GO:0042825) |
| 3.4 | 20.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 3.1 | 9.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.5 | 17.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.4 | 7.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.8 | 16.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 1.6 | 6.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.6 | 11.2 | GO:0001652 | granular component(GO:0001652) |
| 1.6 | 7.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.5 | 8.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.4 | 4.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.4 | 4.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 1.4 | 9.7 | GO:0060205 | cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 1.4 | 6.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 1.3 | 4.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.3 | 4.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.3 | 11.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.3 | 3.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.2 | 5.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.2 | 7.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 1.2 | 10.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 1.1 | 6.8 | GO:0097422 | tubular endosome(GO:0097422) |
| 1.1 | 6.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.1 | 4.5 | GO:0071008 | B cell receptor complex(GO:0019815) U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 1.1 | 4.3 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 1.0 | 11.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.0 | 15.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.9 | 9.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.9 | 9.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.9 | 2.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.9 | 10.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.8 | 6.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.8 | 5.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.8 | 9.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.8 | 4.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.8 | 5.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.7 | 3.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.7 | 14.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.7 | 26.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.7 | 4.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.7 | 4.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.7 | 7.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.6 | 2.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.6 | 1.9 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.6 | 9.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 1.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.5 | 9.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.5 | 12.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 8.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 12.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.5 | 5.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.5 | 9.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.5 | 6.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.5 | 2.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.5 | 2.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.5 | 4.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.5 | 4.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 139.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 5.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 10.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 5.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 1.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.4 | 13.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.4 | 8.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 5.2 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.4 | 1.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.4 | 2.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 9.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 5.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 9.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 1.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.4 | 25.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.4 | 4.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 3.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 5.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 27.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 23.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 2.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 38.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 1.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.3 | 3.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 10.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 3.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 2.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 5.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.3 | 4.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 8.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 2.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.3 | 3.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 3.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 3.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 9.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 2.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 12.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 27.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 31.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.2 | 1.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 0.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 15.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 2.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 6.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 5.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 4.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 2.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.2 | 3.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 6.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 8.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 1.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 2.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 4.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 20.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 10.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 1.2 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 32.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 2.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 4.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 8.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 2.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 3.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 4.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 16.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 5.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 20.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 4.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 3.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 3.2 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 6.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 5.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 4.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.1 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 4.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 12.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 13.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 19.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 35.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 5.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.8 | 78.8 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 10.0 | 29.9 | GO:0032810 | sterol response element binding(GO:0032810) |
| 6.6 | 26.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 5.9 | 17.8 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 4.9 | 24.4 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 4.4 | 21.8 | GO:0004103 | choline kinase activity(GO:0004103) |
| 3.9 | 11.7 | GO:0070976 | TIR domain binding(GO:0070976) |
| 3.7 | 11.1 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 3.7 | 14.7 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 3.7 | 11.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 3.5 | 17.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 3.4 | 13.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 3.3 | 10.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 3.3 | 9.8 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 2.9 | 11.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 2.6 | 18.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 2.6 | 10.5 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.5 | 10.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 2.4 | 9.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 2.4 | 11.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 2.3 | 6.8 | GO:0015292 | uniporter activity(GO:0015292) |
| 2.2 | 17.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 2.2 | 8.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 2.1 | 18.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 2.1 | 6.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 2.1 | 8.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 2.0 | 4.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 2.0 | 14.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 2.0 | 14.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 2.0 | 24.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 2.0 | 6.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 2.0 | 13.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 2.0 | 5.9 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 1.9 | 7.8 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 1.9 | 13.4 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 1.9 | 5.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.9 | 11.4 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 1.9 | 5.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.9 | 5.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.8 | 5.5 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.8 | 8.8 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.8 | 15.9 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.7 | 13.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.7 | 6.9 | GO:0019002 | GMP binding(GO:0019002) |
| 1.7 | 6.7 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 1.7 | 6.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.7 | 11.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 1.6 | 4.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.6 | 11.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.6 | 19.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 1.6 | 12.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.6 | 4.8 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 1.6 | 6.2 | GO:0070401 | NADP+ binding(GO:0070401) estradiol binding(GO:1903924) |
| 1.6 | 9.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.5 | 4.6 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.4 | 4.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 1.4 | 20.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.4 | 5.5 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 1.4 | 4.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 1.4 | 12.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.3 | 4.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 1.3 | 28.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.3 | 26.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.3 | 17.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 1.2 | 3.7 | GO:0005503 | all-trans retinal binding(GO:0005503) benzaldehyde dehydrogenase activity(GO:0019115) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 1.2 | 5.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.2 | 16.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 1.2 | 11.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.2 | 3.5 | GO:0019961 | interferon binding(GO:0019961) |
| 1.2 | 8.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.1 | 6.9 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.1 | 4.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 1.1 | 19.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 1.0 | 5.1 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 1.0 | 9.0 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 1.0 | 3.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.0 | 10.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 1.0 | 4.9 | GO:0001007 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 1.0 | 19.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.9 | 3.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.9 | 3.7 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.9 | 5.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.9 | 3.6 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.9 | 2.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.9 | 5.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.9 | 11.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.9 | 2.7 | GO:0070905 | serine binding(GO:0070905) |
| 0.9 | 10.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.8 | 2.5 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.8 | 2.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.8 | 3.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.8 | 0.8 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.8 | 16.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.8 | 22.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.8 | 4.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) GPI anchor binding(GO:0034235) |
| 0.8 | 6.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.7 | 3.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.7 | 10.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 1.5 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.7 | 3.7 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.7 | 4.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.7 | 4.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.7 | 6.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.7 | 4.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.7 | 15.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.6 | 2.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 10.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.6 | 5.7 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.6 | 6.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.6 | 5.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.6 | 7.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.6 | 12.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.6 | 9.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.6 | 9.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.6 | 4.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.6 | 11.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 1.8 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.6 | 1.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.6 | 2.3 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.6 | 10.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.6 | 2.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 3.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.5 | 2.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 7.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.5 | 3.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 4.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.5 | 4.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.5 | 2.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.5 | 1.6 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.5 | 16.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.5 | 11.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.5 | 1.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.5 | 5.1 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.5 | 5.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.5 | 6.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 6.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.5 | 2.9 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.5 | 5.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 12.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 3.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.5 | 6.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.5 | 2.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.5 | 4.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 11.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.4 | 5.4 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.4 | 5.7 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of synapse(GO:0098918) structural constituent of postsynaptic density(GO:0098919) |
| 0.4 | 1.7 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.4 | 8.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.4 | 10.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 1.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.4 | 1.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 22.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.4 | 27.3 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.4 | 5.9 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.4 | 8.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 4.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 1.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.4 | 8.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 4.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 8.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 11.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 6.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 6.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.3 | 2.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.3 | 1.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 9.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 3.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.3 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 1.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 3.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 5.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 12.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.3 | 1.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.3 | 5.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 9.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.3 | 20.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.3 | 35.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.3 | 7.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 5.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 4.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 2.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 6.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 6.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 7.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 4.7 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.2 | 10.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 13.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.2 | 2.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.2 | 6.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 1.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 2.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 7.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.2 | 9.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 28.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 13.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.2 | 5.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 3.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 9.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 1.5 | GO:0060229 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.2 | 4.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 6.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 8.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 6.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.2 | 19.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 23.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 14.1 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 5.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 2.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 2.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.9 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 3.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.6 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 1.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.8 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 4.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 10.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 2.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 2.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 18.0 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 4.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 3.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 2.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 15.9 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 15.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 4.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.6 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 9.2 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 5.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0004177 | aminopeptidase activity(GO:0004177) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 68.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.9 | 123.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 1.5 | 33.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.9 | 22.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.9 | 13.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.8 | 25.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.8 | 14.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.8 | 9.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.7 | 23.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.7 | 29.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.7 | 17.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.6 | 13.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.6 | 10.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.6 | 17.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.6 | 3.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.6 | 8.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 14.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.4 | 10.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.4 | 7.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 15.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.4 | 17.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 6.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 1.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 4.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 7.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.3 | 14.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 2.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 9.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 23.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.3 | 18.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.3 | 11.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 8.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 2.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 5.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 9.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 4.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 2.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 3.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 6.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 4.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 5.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 56.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 3.5 | 157.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 3.1 | 18.8 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 2.4 | 19.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 1.9 | 13.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 1.9 | 44.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 1.6 | 21.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 1.5 | 10.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 1.3 | 11.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.2 | 20.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.1 | 16.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.0 | 34.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 1.0 | 13.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.9 | 31.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.9 | 11.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.9 | 20.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.9 | 9.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.9 | 16.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.8 | 11.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.8 | 6.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.8 | 7.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.8 | 13.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.8 | 4.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.7 | 11.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.7 | 12.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.7 | 23.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.7 | 8.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.6 | 11.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.6 | 35.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.6 | 10.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.6 | 6.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.6 | 16.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.6 | 13.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.6 | 3.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.6 | 16.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.6 | 7.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.5 | 22.7 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.5 | 18.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.5 | 14.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 19.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 8.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 12.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 6.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.5 | 5.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 9.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 21.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 8.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.4 | 6.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 4.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 3.7 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 6.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 9.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.4 | 24.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 4.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 10.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.3 | 8.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.3 | 12.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 10.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.3 | 3.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.3 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 15.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 4.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 7.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 5.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 17.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 4.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 9.0 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.2 | 3.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 5.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 3.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 0.9 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 2.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 2.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 4.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 23.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 10.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 9.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.2 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 17.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 5.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 3.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 5.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 4.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 2.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |