Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
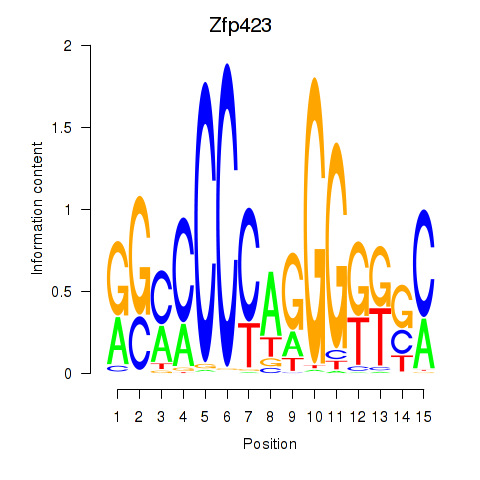
Results for Zfp423
Z-value: 0.44
Transcription factors associated with Zfp423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp423
|
ENSRNOG00000014658 | zinc finger protein 423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp423 | rn6_v1_chr19_+_20147037_20147037 | 0.25 | 4.3e-06 | Click! |
Activity profile of Zfp423 motif
Sorted Z-values of Zfp423 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_142453013 | 21.13 |
ENSRNOT00000056573
|
AABR07061755.1
|
|
| chr1_-_226152524 | 15.90 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr6_-_39363367 | 13.60 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr1_-_70235091 | 12.54 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr3_+_56868167 | 9.93 |
ENSRNOT00000087134
|
Gad1
|
glutamate decarboxylase 1 |
| chr13_-_90710148 | 9.56 |
ENSRNOT00000010113
ENSRNOT00000080638 |
Kcnj9
|
potassium voltage-gated channel subfamily J member 9 |
| chr12_-_47793534 | 8.50 |
ENSRNOT00000001588
|
Fam222a
|
family with sequence similarity 222, member A |
| chr1_-_226924244 | 7.33 |
ENSRNOT00000028971
|
Tmem132a
|
transmembrane protein 132A |
| chr7_-_107009330 | 6.83 |
ENSRNOT00000074573
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr15_-_28733513 | 6.17 |
ENSRNOT00000078180
|
Sall2
|
spalt-like transcription factor 2 |
| chr1_+_224800252 | 6.11 |
ENSRNOT00000024488
|
Slc22a8
|
solute carrier family 22 member 8 |
| chr7_-_124041594 | 5.95 |
ENSRNOT00000064700
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr10_+_80790168 | 5.74 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr1_+_87248489 | 5.72 |
ENSRNOT00000028091
|
Dpf1
|
double PHD fingers 1 |
| chr6_-_44361908 | 5.31 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr1_+_100224401 | 4.88 |
ENSRNOT00000025846
|
Fv1
|
Friend virus susceptibility 1 |
| chr1_-_170464341 | 4.10 |
ENSRNOT00000024850
|
Trim3
|
tripartite motif-containing 3 |
| chr9_+_117538346 | 4.01 |
ENSRNOT00000022849
|
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr7_+_120067379 | 3.96 |
ENSRNOT00000011270
|
Cdc42ep1
|
CDC42 effector protein 1 |
| chr1_+_101688297 | 3.80 |
ENSRNOT00000091606
|
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr4_+_7282948 | 3.40 |
ENSRNOT00000011052
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr1_+_266953139 | 3.35 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr4_+_32373641 | 2.76 |
ENSRNOT00000076086
|
Dlx6
|
distal-less homeobox 6 |
| chr5_+_172648950 | 2.74 |
ENSRNOT00000055361
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr1_-_131454689 | 2.71 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_-_70926903 | 2.68 |
ENSRNOT00000031005
|
Lrp1
|
LDL receptor related protein 1 |
| chr4_-_157009674 | 2.55 |
ENSRNOT00000071625
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr4_-_156427755 | 2.55 |
ENSRNOT00000085675
ENSRNOT00000014030 |
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr4_-_157008947 | 2.47 |
ENSRNOT00000073341
|
Pex5
|
peroxisomal biogenesis factor 5 |
| chr2_-_48501436 | 2.40 |
ENSRNOT00000017305
|
Isl1
|
ISL LIM homeobox 1 |
| chr11_+_86890585 | 1.83 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr1_-_259484569 | 1.81 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_-_198316882 | 1.51 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr19_+_43163129 | 1.40 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr3_+_161252723 | 1.29 |
ENSRNOT00000084283
|
Snx21
|
sorting nexin family member 21 |
| chr5_+_138279506 | 0.98 |
ENSRNOT00000084651
|
P3h1
|
prolyl 3-hydroxylase 1 |
| chr13_+_51384562 | 0.97 |
ENSRNOT00000077928
|
Kdm5b
|
lysine demethylase 5B |
| chr2_-_61949926 | 0.96 |
ENSRNOT00000025966
|
Npr3
|
natriuretic peptide receptor 3 |
| chr7_+_130532435 | 0.92 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr10_-_18131562 | 0.86 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chrX_+_140878216 | 0.84 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr8_+_82038967 | 0.70 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr17_-_21353134 | 0.68 |
ENSRNOT00000067898
|
Smim13
|
small integral membrane protein 13 |
| chr5_-_138279285 | 0.66 |
ENSRNOT00000009044
|
RGD1564804
|
similar to chromosome 1 open reading frame 50 |
| chr9_+_94178221 | 0.63 |
ENSRNOT00000033487
|
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr13_+_51384389 | 0.62 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr3_-_122920684 | 0.61 |
ENSRNOT00000028818
|
Cpxm1
|
carboxypeptidase X (M14 family), member 1 |
| chr7_-_144960527 | 0.56 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr19_-_11057254 | 0.41 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr2_-_210413985 | 0.30 |
ENSRNOT00000025036
|
Strip1
|
striatin interacting protein 1 |
| chr11_-_86890390 | 0.29 |
ENSRNOT00000046183
|
Trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr5_-_171312026 | 0.25 |
ENSRNOT00000063779
|
Smim1
|
small integral membrane protein 1 |
| chr1_-_8878136 | 0.04 |
ENSRNOT00000064836
ENSRNOT00000075850 |
Vta1
|
vesicle trafficking 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp423
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 2.0 | 9.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.8 | 5.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 1.1 | 7.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.9 | 15.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.9 | 2.7 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.8 | 2.4 | GO:0071657 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.7 | 2.7 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.7 | 4.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.6 | 1.8 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.4 | 1.6 | GO:2000864 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.4 | 3.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.4 | 1.8 | GO:1904008 | response to monosodium glutamate(GO:1904008) |
| 0.3 | 1.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.3 | 6.8 | GO:0061548 | membrane hyperpolarization(GO:0060081) ganglion development(GO:0061548) |
| 0.3 | 9.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 4.0 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 4.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 2.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.7 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.1 | 0.6 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 5.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 1.0 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.4 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.9 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 2.8 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 1.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 6.2 | GO:0001654 | eye development(GO:0001654) |
| 0.0 | 3.8 | GO:0001889 | liver development(GO:0001889) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.6 | 1.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 5.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 6.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 4.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 6.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.3 | 9.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.7 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.2 | 2.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 2.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 5.0 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 5.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 4.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 6.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 18.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 10.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 2.0 | 9.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.9 | 9.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.5 | 7.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.0 | 5.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.8 | 6.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.7 | 3.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.3 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.3 | 1.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 4.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 2.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 3.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 11.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 4.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 4.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.8 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 15.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.6 | 6.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 9.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 9.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 6.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 5.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 3.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.8 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |