Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
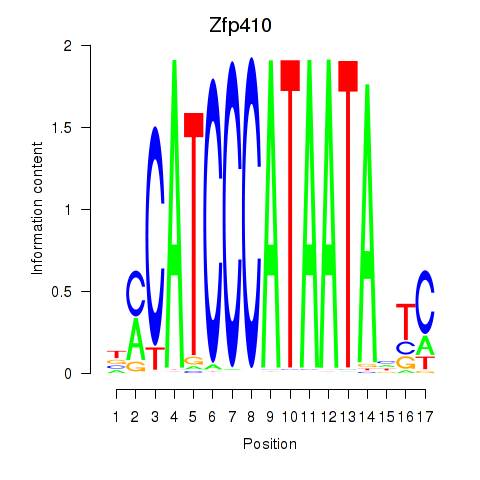
Results for Zfp410
Z-value: 0.30
Transcription factors associated with Zfp410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp410
|
ENSRNOG00000010985 | zinc finger protein 410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp410 | rn6_v1_chr6_+_108032856_108032856 | -0.05 | 3.4e-01 | Click! |
Activity profile of Zfp410 motif
Sorted Z-values of Zfp410 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_157612536 | 39.85 |
ENSRNOT00000055970
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr15_-_41595345 | 12.22 |
ENSRNOT00000019639
|
Sgcg
|
sarcoglycan, gamma |
| chr18_-_61707307 | 3.91 |
ENSRNOT00000085784
ENSRNOT00000032047 |
Lman1
|
lectin, mannose-binding, 1 |
| chr5_+_132005738 | 3.55 |
ENSRNOT00000041421
|
Skint4
|
selection and upkeep of intraepithelial T cells 4 |
| chr6_-_26138414 | 3.35 |
ENSRNOT00000034712
|
Mrpl33
|
mitochondrial ribosomal protein L33 |
| chr7_+_12941994 | 2.55 |
ENSRNOT00000010832
|
Odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr2_-_205451391 | 2.51 |
ENSRNOT00000036229
|
Nr1h5
|
nuclear receptor subfamily 1, group H, member 5 |
| chr10_-_108150511 | 2.39 |
ENSRNOT00000073337
|
Cbx8
|
chromobox 8 |
| chr12_-_47510263 | 1.31 |
ENSRNOT00000034129
|
LOC691311
|
hypothetical protein LOC691311 |
| chr1_-_66212418 | 0.86 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr5_-_169331163 | 0.80 |
ENSRNOT00000042301
|
Espn
|
espin |
| chr9_+_17817721 | 0.71 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr1_-_274106502 | 0.67 |
ENSRNOT00000020539
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr10_-_31712924 | 0.51 |
ENSRNOT00000067312
|
Dppa1
|
developmental pluripotency associated 1 |
| chr10_+_34804878 | 0.12 |
ENSRNOT00000043954
|
Olr1401
|
olfactory receptor 1401 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp410
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 39.9 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 3.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.7 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.1 | 2.4 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 12.1 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 2.5 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.7 | 39.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 3.9 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.2 | 2.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 3.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 39.9 | GO:0031492 | RNA polymerase II repressing transcription factor binding(GO:0001103) nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.7 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.2 | 2.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 3.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 3.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 39.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.9 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |