Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
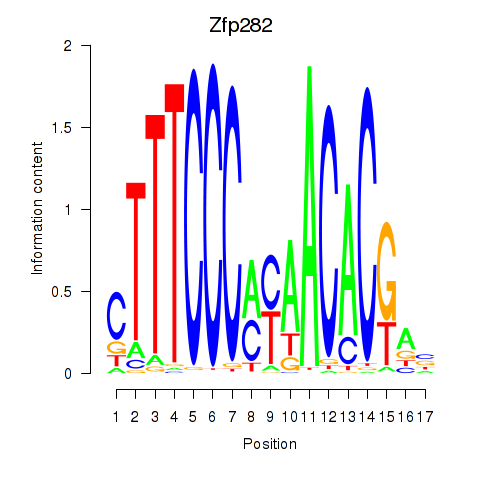
Results for Zfp282
Z-value: 0.51
Transcription factors associated with Zfp282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp282
|
ENSRNOG00000026981 | zinc finger protein 282 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp282 | rn6_v1_chr4_+_77554269_77554269 | 0.05 | 3.9e-01 | Click! |
Activity profile of Zfp282 motif
Sorted Z-values of Zfp282 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_65566171 | 16.95 |
ENSRNOT00000015651
|
Spag5
|
sperm associated antigen 5 |
| chr16_-_48921242 | 15.97 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chr7_+_140758615 | 14.20 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr2_-_165641573 | 13.28 |
ENSRNOT00000013987
|
Trim59
|
tripartite motif-containing 59 |
| chr20_-_3397039 | 12.45 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_157700064 | 12.42 |
ENSRNOT00000031974
|
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr4_+_78320190 | 11.79 |
ENSRNOT00000032742
ENSRNOT00000091359 |
Gimap4
|
GTPase, IMAP family member 4 |
| chr7_-_61798729 | 10.46 |
ENSRNOT00000010283
|
Dyrk2
|
dual specificity tyrosine phosphorylation regulated kinase 2 |
| chr2_+_190073815 | 10.27 |
ENSRNOT00000015473
|
S100a8
|
S100 calcium binding protein A8 |
| chr18_+_87415531 | 9.27 |
ENSRNOT00000072475
|
AABR07032888.1
|
|
| chrX_-_157848842 | 9.15 |
ENSRNOT00000035398
|
Fam122c
|
family with sequence similarity 122C |
| chr2_+_187447501 | 8.95 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr5_+_154522119 | 7.56 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr4_+_30313102 | 7.27 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr10_+_16542882 | 7.21 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr1_-_220867815 | 7.14 |
ENSRNOT00000028015
|
Mus81
|
MUS81 structure-specific endonuclease subunit |
| chr10_-_90312386 | 7.07 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr14_-_44375804 | 6.28 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr15_-_34693034 | 6.06 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr9_+_10760113 | 5.43 |
ENSRNOT00000073054
|
Arrdc5
|
arrestin domain containing 5 |
| chr3_-_132282879 | 5.31 |
ENSRNOT00000042040
|
LOC100361388
|
high-mobility group nucleosomal binding domain 2-like |
| chr5_+_162127810 | 5.04 |
ENSRNOT00000038858
|
Pramef27
|
PRAME family member 27 |
| chr15_+_12570062 | 4.97 |
ENSRNOT00000010675
|
Thoc7
|
THO complex 7 |
| chr14_+_113968563 | 4.95 |
ENSRNOT00000006085
|
Clhc1
|
clathrin heavy chain linker domain containing 1 |
| chrX_+_78042859 | 4.73 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr20_-_46871946 | 4.61 |
ENSRNOT00000031047
|
Armc2
|
armadillo repeat containing 2 |
| chr7_-_60743328 | 4.56 |
ENSRNOT00000066767
|
Mdm2
|
MDM2 proto-oncogene |
| chr17_-_66667853 | 4.34 |
ENSRNOT00000058749
|
A26c2
|
ANKRD26-like family C, member 2 |
| chr8_-_48564722 | 4.26 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr1_+_97712177 | 4.22 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr1_+_72380711 | 4.16 |
ENSRNOT00000022236
|
Fiz1
|
FLT3-interacting zinc finger 1 |
| chr8_-_90664554 | 4.06 |
ENSRNOT00000039917
ENSRNOT00000074896 ENSRNOT00000072225 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr7_-_11777503 | 3.87 |
ENSRNOT00000026220
|
Amh
|
anti-Mullerian hormone |
| chr2_-_189307108 | 3.62 |
ENSRNOT00000028246
|
Atp8b2
|
ATPase phospholipid transporting 8B2 |
| chr2_+_240527130 | 3.42 |
ENSRNOT00000031348
|
Slc9b1
|
solute carrier family 9 member B1 |
| chr18_-_786674 | 3.41 |
ENSRNOT00000021955
|
Cetn1
|
centrin 1 |
| chr2_+_27365148 | 3.11 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr3_-_14643897 | 3.01 |
ENSRNOT00000082008
ENSRNOT00000025983 |
Ggta1
|
glycoprotein, alpha-galactosyltransferase 1 |
| chr9_-_110936631 | 2.82 |
ENSRNOT00000064431
|
Fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chrX_+_96667863 | 2.68 |
ENSRNOT00000042552
|
RGD1561151
|
similar to hypothetical protein 4932411N23 |
| chr1_+_266953139 | 2.59 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr3_-_151224123 | 2.52 |
ENSRNOT00000026091
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr4_+_118167294 | 2.45 |
ENSRNOT00000022367
|
LOC687679
|
similar to small nuclear ribonucleoprotein polypeptide G |
| chr1_-_169001761 | 2.32 |
ENSRNOT00000048484
|
Hbe2
|
hemoglobin, epsilon 2 |
| chr8_+_117694605 | 2.23 |
ENSRNOT00000027994
|
Col7a1
|
collagen type VII alpha 1 chain |
| chr1_+_167179452 | 2.22 |
ENSRNOT00000045726
|
LOC102549471
|
short transient receptor potential channel 2-like |
| chr14_+_44524416 | 2.21 |
ENSRNOT00000038927
|
Rpl9
|
ribosomal protein L9 |
| chr1_+_85213652 | 2.16 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr14_+_44524753 | 2.15 |
ENSRNOT00000076245
|
Rpl9
|
ribosomal protein L9 |
| chrX_-_57058526 | 2.04 |
ENSRNOT00000091191
|
AABR07038624.1
|
|
| chr8_+_50563047 | 1.98 |
ENSRNOT00000025149
|
Zfp259
|
zinc finger protein 259 |
| chr1_-_79259492 | 1.97 |
ENSRNOT00000043333
|
Psg29
|
pregnancy-specific glycoprotein 29 |
| chr2_+_248276709 | 1.90 |
ENSRNOT00000068683
|
Gbp2
|
guanylate binding protein 2 |
| chr4_+_66091641 | 1.88 |
ENSRNOT00000043147
|
AABR07060287.1
|
|
| chr3_+_103753238 | 1.68 |
ENSRNOT00000007144
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr10_+_14828597 | 1.59 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chrX_+_115563038 | 1.58 |
ENSRNOT00000087859
|
Alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr1_-_154111725 | 1.52 |
ENSRNOT00000055488
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr9_-_4447715 | 1.52 |
ENSRNOT00000061882
|
Kat2b
|
lysine acetyltransferase 2B |
| chr8_-_42365819 | 1.35 |
ENSRNOT00000071125
|
LOC103692018
|
olfactory receptor 8B3-like |
| chr1_+_172099992 | 1.27 |
ENSRNOT00000049763
|
Olr240
|
olfactory receptor 240 |
| chr18_+_40962146 | 1.25 |
ENSRNOT00000035656
|
Arl14epl
|
ADP ribosylation factor like GTPase 14 effector protein like |
| chr1_-_75354820 | 1.21 |
ENSRNOT00000018989
|
RGD1310257
|
similar to RIKEN cDNA 6330408A02 gene |
| chr17_-_78499881 | 1.13 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chr1_-_86948845 | 1.09 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chr3_-_104504204 | 1.02 |
ENSRNOT00000049582
|
Ryr3
|
ryanodine receptor 3 |
| chr1_+_187149453 | 0.97 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr4_-_1686845 | 0.91 |
ENSRNOT00000073139
|
LOC100912507
|
olfactory receptor 8B3-like |
| chr5_-_150653172 | 0.87 |
ENSRNOT00000072028
|
Med18
|
mediator complex subunit 18 |
| chr8_-_68312909 | 0.86 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr14_-_84662143 | 0.70 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr6_-_21880003 | 0.66 |
ENSRNOT00000006505
|
Ttc27
|
tetratricopeptide repeat domain 27 |
| chr17_-_5463158 | 0.60 |
ENSRNOT00000090032
|
Naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr8_-_54994871 | 0.60 |
ENSRNOT00000013014
|
Tex12
|
testis expressed 12 |
| chr7_-_5106708 | 0.52 |
ENSRNOT00000046001
|
Olr892
|
olfactory receptor 892 |
| chrX_-_1786978 | 0.52 |
ENSRNOT00000011317
|
Rbm10
|
RNA binding motif protein 10 |
| chr19_+_38601786 | 0.47 |
ENSRNOT00000087655
|
Zfp90
|
zinc finger protein 90 |
| chr1_+_172129514 | 0.45 |
ENSRNOT00000041658
|
Olr241
|
olfactory receptor 241 |
| chr10_-_44243402 | 0.43 |
ENSRNOT00000058517
|
LOC691352
|
similar to Robo-1 |
| chr12_-_43503456 | 0.39 |
ENSRNOT00000078445
|
Med13l
|
mediator complex subunit 13-like |
| chr8_-_37093082 | 0.32 |
ENSRNOT00000072767
|
Pate-f
|
prostate and testis expressed protein F |
| chr20_+_29558689 | 0.28 |
ENSRNOT00000085229
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr1_-_197982601 | 0.25 |
ENSRNOT00000025782
|
Eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr8_-_118926613 | 0.15 |
ENSRNOT00000056130
|
Nbeal2
|
neurobeachin-like 2 |
| chr1_-_7443863 | 0.13 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr9_+_99720856 | 0.09 |
ENSRNOT00000047487
|
Olr1346
|
olfactory receptor 1346 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp282
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.5 | 4.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) response to formaldehyde(GO:1904404) |
| 1.4 | 7.1 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 1.3 | 3.9 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 1.3 | 7.6 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.2 | 12.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 1.2 | 10.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 1.0 | 16.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.0 | 3.0 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.7 | 9.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.6 | 3.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.6 | 10.3 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.6 | 13.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.6 | 1.7 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 7.3 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.5 | 1.5 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.5 | 2.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) Cajal body organization(GO:0030576) |
| 0.5 | 7.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 5.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.4 | 4.3 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 1.6 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.2 | 2.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 10.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 1.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 2.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 4.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.9 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 3.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.5 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 1.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.7 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 2.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 4.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 3.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 2.2 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 3.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.0 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.5 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 9.6 | GO:0043009 | chordate embryonic development(GO:0043009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.9 | 7.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.5 | 17.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.5 | 5.0 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.5 | 13.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.5 | 1.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.4 | 1.6 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 4.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 2.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 2.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 16.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 2.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 2.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.2 | 5.0 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 9.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 7.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 10.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 1.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 10.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.6 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 6.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 7.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.5 | 9.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.2 | 4.7 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 7.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.8 | 4.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.8 | 3.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.7 | 3.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 3.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 1.7 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 7.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.3 | 1.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.3 | 1.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.3 | 4.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 3.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 10.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 3.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 2.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 2.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 24.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 6.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 11.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 4.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 15.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 7.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 13.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 6.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 6.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 11.7 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.4 | 10.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 3.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 4.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 12.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 13.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.2 | 4.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 9.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.1 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 4.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 4.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 4.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 8.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 4.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 7.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |