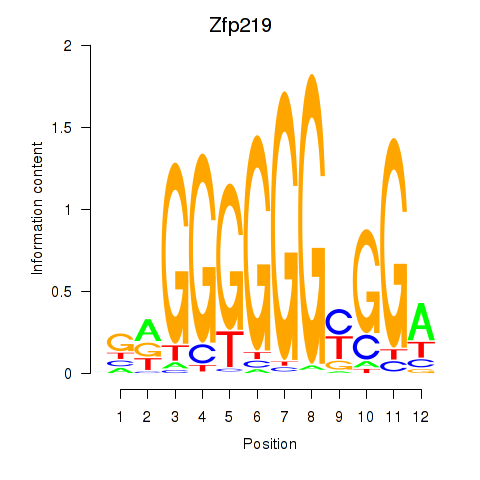
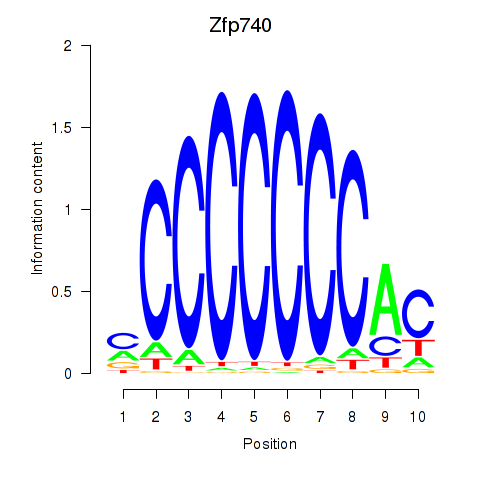
Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zfp219_Zfp740
Z-value: 0.59
Transcription factors associated with Zfp219_Zfp740
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp219
|
ENSRNOG00000011544 | zinc finger protein 219 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp219 | rn6_v1_chr15_-_28406046_28406046 | 0.13 | 1.7e-02 | Click! |
| Znf740 | rn6_v1_chr7_+_143810892_143810892 | -0.06 | 3.0e-01 | Click! |
Activity profile of Zfp219_Zfp740 motif
Sorted Z-values of Zfp219_Zfp740 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_24631679 | 15.94 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr3_-_23020441 | 14.06 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr20_+_4959294 | 10.64 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr11_+_60253608 | 9.29 |
ENSRNOT00000090804
|
LOC685716
|
similar to OX-2 membrane glycoprotein precursor (MRC OX-2 antigen) (CD200 antigen) |
| chr6_-_146195819 | 9.04 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr20_+_8696436 | 8.53 |
ENSRNOT00000085131
|
Zfand3
|
zinc finger AN1-type containing 3 |
| chr4_-_157750088 | 8.28 |
ENSRNOT00000038023
|
Cd27
|
CD27 molecule |
| chr10_-_64862268 | 8.18 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr1_+_85112834 | 7.61 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr3_+_62481323 | 7.60 |
ENSRNOT00000078872
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr2_-_189400323 | 6.98 |
ENSRNOT00000024364
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chr2_+_43266739 | 6.79 |
ENSRNOT00000087031
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr10_-_89084885 | 6.66 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chr4_+_157612536 | 6.45 |
ENSRNOT00000055970
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr7_+_144052061 | 6.31 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr2_+_201289357 | 6.07 |
ENSRNOT00000067358
|
Tbx15
|
T-box 15 |
| chr7_+_144014173 | 6.07 |
ENSRNOT00000019403
|
Sp1
|
Sp1 transcription factor |
| chr10_+_91217079 | 5.84 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr18_+_27576129 | 5.82 |
ENSRNOT00000070930
|
Kdm3b
|
lysine demethylase 3B |
| chr5_-_59025631 | 5.73 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr2_+_195996521 | 5.64 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr17_+_49516429 | 5.58 |
ENSRNOT00000048929
|
Tgif2-ps1
|
TGFB-induced factor homeobox 2, pseudogene 1 |
| chr20_+_4355175 | 5.28 |
ENSRNOT00000000510
|
Gpsm3
|
G-protein signaling modulator 3 |
| chr5_+_151211342 | 5.15 |
ENSRNOT00000067939
|
Ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr1_+_243477493 | 5.10 |
ENSRNOT00000021779
ENSRNOT00000085356 |
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr5_-_152247332 | 5.06 |
ENSRNOT00000077165
|
Lin28a
|
lin-28 homolog A |
| chr13_+_52976507 | 5.01 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr7_-_143863186 | 4.95 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr6_-_12554439 | 4.93 |
ENSRNOT00000022481
|
Lhcgr
|
luteinizing hormone/choriogonadotropin receptor |
| chr3_+_33641616 | 4.74 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr9_-_92435363 | 4.66 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chrX_+_71155601 | 4.63 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr7_-_70969905 | 4.53 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr18_-_15167311 | 4.39 |
ENSRNOT00000021061
|
Rnf138
|
ring finger protein 138 |
| chr9_-_61974898 | 4.38 |
ENSRNOT00000091519
|
Boll
|
boule homolog, RNA binding protein |
| chr2_+_113984646 | 4.33 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr2_+_198040536 | 4.32 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr1_-_127599257 | 4.27 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr9_-_64573076 | 4.26 |
ENSRNOT00000084658
|
LOC108348134
|
protein boule-like |
| chr12_-_9331195 | 4.21 |
ENSRNOT00000044134
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chrX_+_109940350 | 4.20 |
ENSRNOT00000093543
ENSRNOT00000057049 |
Nrk
|
Nik related kinase |
| chr13_+_89524329 | 4.18 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr8_-_23084879 | 4.17 |
ENSRNOT00000018874
|
Zfp653
|
zinc finger protein 653 |
| chr20_+_4357733 | 4.12 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr4_-_152835182 | 4.12 |
ENSRNOT00000036721
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr5_-_151824633 | 4.01 |
ENSRNOT00000043959
|
Sfn
|
stratifin |
| chr1_+_198932870 | 3.98 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr1_+_199225100 | 3.98 |
ENSRNOT00000088606
|
Setd1a
|
SET domain containing 1A |
| chr10_-_90127600 | 3.97 |
ENSRNOT00000028368
|
Lsm12
|
LSM12 homolog |
| chr1_-_197919480 | 3.94 |
ENSRNOT00000068529
ENSRNOT00000080605 |
Atxn2l
|
ataxin 2-like |
| chr4_-_77347011 | 3.93 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr7_+_71004417 | 3.78 |
ENSRNOT00000005575
|
Myo1a
|
myosin IA |
| chr8_+_22648323 | 3.71 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr4_-_56493923 | 3.67 |
ENSRNOT00000027173
|
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr6_+_23757225 | 3.65 |
ENSRNOT00000005553
|
LOC298795
|
similar to 14-3-3 protein sigma |
| chr10_-_89338739 | 3.65 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr3_+_172420591 | 3.62 |
ENSRNOT00000071215
|
Gnas
|
GNAS complex locus |
| chr8_+_59344083 | 3.58 |
ENSRNOT00000031175
|
Crabp1
|
cellular retinoic acid binding protein 1 |
| chr6_-_22281724 | 3.58 |
ENSRNOT00000079137
|
Spast
|
spastin |
| chr17_-_20364714 | 3.49 |
ENSRNOT00000070962
|
Jarid2
|
jumonji and AT-rich interaction domain containing 2 |
| chr5_-_147375009 | 3.44 |
ENSRNOT00000009436
|
S100pbp
|
S100P binding protein |
| chr6_+_137184820 | 3.44 |
ENSRNOT00000073796
|
Adssl1
|
adenylosuccinate synthase like 1 |
| chr1_-_154216340 | 3.43 |
ENSRNOT00000024082
|
Eed
|
embryonic ectoderm development |
| chr10_+_34519790 | 3.33 |
ENSRNOT00000052360
|
Mgat1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr2_-_188660179 | 3.30 |
ENSRNOT00000027935
|
Efna4
|
ephrin A4 |
| chr17_-_76287186 | 3.21 |
ENSRNOT00000092452
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr3_-_112789282 | 3.20 |
ENSRNOT00000090680
ENSRNOT00000066181 |
Ttbk2
|
tau tubulin kinase 2 |
| chr12_-_21760292 | 3.19 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr10_+_56610051 | 3.17 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr7_-_75422268 | 3.13 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr13_+_36532758 | 3.11 |
ENSRNOT00000083007
|
En1
|
engrailed 1 |
| chr11_-_83926524 | 3.01 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_-_22281886 | 2.93 |
ENSRNOT00000039375
|
Spast
|
spastin |
| chr10_+_11206226 | 2.89 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr3_-_156340913 | 2.86 |
ENSRNOT00000021452
|
Mafb
|
MAF bZIP transcription factor B |
| chr8_-_128521109 | 2.82 |
ENSRNOT00000034025
|
Scn11a
|
sodium voltage-gated channel alpha subunit 11 |
| chr10_+_74910713 | 2.81 |
ENSRNOT00000091619
|
Hsf5
|
heat shock transcription factor 5 |
| chr4_-_99546905 | 2.80 |
ENSRNOT00000077447
|
Kdm3a
|
lysine demethylase 3A |
| chr8_+_118821729 | 2.79 |
ENSRNOT00000028409
|
Setd2
|
SET domain containing 2 |
| chr5_-_169167831 | 2.79 |
ENSRNOT00000012407
|
Phf13
|
PHD finger protein 13 |
| chr11_-_61499557 | 2.65 |
ENSRNOT00000046208
|
Usf3
|
upstream transcription factor family member 3 |
| chr20_-_7930929 | 2.60 |
ENSRNOT00000000607
|
Tead3
|
TEA domain transcription factor 3 |
| chr20_+_5125679 | 2.59 |
ENSRNOT00000060832
|
Bag6
|
BCL2-associated athanogene 6 |
| chr6_-_95998529 | 2.58 |
ENSRNOT00000050138
|
Six4
|
SIX homeobox 4 |
| chr10_-_76039964 | 2.54 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_+_144078496 | 2.51 |
ENSRNOT00000055302
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr2_+_187512164 | 2.47 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chr5_+_57472315 | 2.47 |
ENSRNOT00000015575
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr7_+_70364813 | 2.45 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr2_+_188253378 | 2.44 |
ENSRNOT00000085690
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr12_+_23151180 | 2.44 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr11_-_61530830 | 2.40 |
ENSRNOT00000059666
ENSRNOT00000068345 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr15_+_34493138 | 2.38 |
ENSRNOT00000089584
ENSRNOT00000027789 |
Nfatc4
|
nuclear factor of activated T-cells 4 |
| chr10_+_61685645 | 2.35 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr10_+_23661013 | 2.34 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr1_+_23977688 | 2.34 |
ENSRNOT00000014805
|
Tbpl1
|
TATA-box binding protein like 1 |
| chr3_+_151126591 | 2.31 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr3_+_120726906 | 2.30 |
ENSRNOT00000051069
|
Bcl2l11
|
BCL2 like 11 |
| chr11_-_61530567 | 2.14 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr15_+_23665202 | 2.08 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr20_+_4572100 | 2.04 |
ENSRNOT00000000476
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr14_+_83341851 | 2.03 |
ENSRNOT00000086090
|
Pisd
|
phosphatidylserine decarboxylase |
| chr10_+_86157608 | 2.03 |
ENSRNOT00000082668
ENSRNOT00000008256 |
Cdk12
|
cyclin-dependent kinase 12 |
| chr10_-_65963932 | 2.02 |
ENSRNOT00000011726
|
Nlk
|
nemo like kinase |
| chr7_-_144966370 | 2.02 |
ENSRNOT00000083935
|
Zfp385a
|
zinc finger protein 385A |
| chr8_-_67869019 | 2.01 |
ENSRNOT00000066009
|
Pias1
|
protein inhibitor of activated STAT, 1 |
| chr1_+_82151669 | 2.01 |
ENSRNOT00000091357
|
Cic
|
capicua transcriptional repressor |
| chr12_-_39882030 | 1.99 |
ENSRNOT00000078761
ENSRNOT00000048851 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr19_-_10358695 | 1.95 |
ENSRNOT00000019770
|
Katnb1
|
katanin regulatory subunit B1 |
| chr20_+_5527181 | 1.93 |
ENSRNOT00000091364
|
Phf1
|
PHD finger protein 1 |
| chr19_+_37600148 | 1.93 |
ENSRNOT00000023853
|
Ctcf
|
CCCTC-binding factor |
| chr7_+_12179203 | 1.90 |
ENSRNOT00000049170
|
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr1_+_40816107 | 1.89 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr20_+_4576057 | 1.88 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr13_+_48607308 | 1.86 |
ENSRNOT00000063882
|
Slc41a1
|
solute carrier family 41 member 1 |
| chr3_-_93734282 | 1.85 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr4_+_98027554 | 1.83 |
ENSRNOT00000009503
|
Serbp1
|
Serpine1 mRNA binding protein 1 |
| chr10_-_108196217 | 1.83 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr20_+_5526975 | 1.79 |
ENSRNOT00000059548
|
Phf1
|
PHD finger protein 1 |
| chr5_-_158313426 | 1.76 |
ENSRNOT00000025488
|
Pax7
|
paired box 7 |
| chr4_-_77706994 | 1.75 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr10_+_93520132 | 1.73 |
ENSRNOT00000055137
|
Mrc2
|
mannose receptor, C type 2 |
| chr1_+_154377447 | 1.72 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_-_34269851 | 1.72 |
ENSRNOT00000026279
|
Psme2
|
proteasome activator subunit 2 |
| chrX_+_33599671 | 1.66 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr19_+_25181564 | 1.63 |
ENSRNOT00000008104
|
Rfx1
|
regulatory factor X1 |
| chr5_-_151898022 | 1.63 |
ENSRNOT00000000133
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr3_+_65815080 | 1.63 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chrX_+_76786466 | 1.59 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chr20_+_4576514 | 1.59 |
ENSRNOT00000090125
ENSRNOT00000047370 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chr7_+_27240876 | 1.59 |
ENSRNOT00000091725
|
LOC362863
|
first gene upstream of Nt5dc3 |
| chr6_-_80334522 | 1.57 |
ENSRNOT00000059316
|
Fbxo33
|
F-box protein 33 |
| chr12_+_22274828 | 1.53 |
ENSRNOT00000001914
|
Epo
|
erythropoietin |
| chr2_+_182796728 | 1.52 |
ENSRNOT00000032351
|
LOC100909434
|
protocadherin-16-like |
| chr6_-_107080524 | 1.52 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr5_+_147375350 | 1.47 |
ENSRNOT00000010674
|
Yars
|
tyrosyl-tRNA synthetase |
| chr2_+_206392200 | 1.46 |
ENSRNOT00000026631
|
Rsbn1
|
round spermatid basic protein 1 |
| chr10_-_84915471 | 1.39 |
ENSRNOT00000087322
|
Sp2
|
Sp2 transcription factor |
| chr1_+_190852985 | 1.38 |
ENSRNOT00000040963
|
Polr3e
|
RNA polymerase III subunit E |
| chr10_+_94407559 | 1.38 |
ENSRNOT00000013046
|
Ddx42
|
DEAD-box helicase 42 |
| chr10_+_84955864 | 1.30 |
ENSRNOT00000038572
|
Sp6
|
Sp6 transcription factor |
| chr20_+_5125349 | 1.29 |
ENSRNOT00000085598
ENSRNOT00000001129 |
Bag6
|
BCL2-associated athanogene 6 |
| chr2_+_209097927 | 1.28 |
ENSRNOT00000023807
|
Dennd2d
|
DENN domain containing 2D |
| chr1_-_72339395 | 1.26 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr3_-_61581598 | 1.24 |
ENSRNOT00000002156
|
Evx2
|
even-skipped homeobox 2 |
| chr8_+_71216178 | 1.18 |
ENSRNOT00000021372
|
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr7_-_140546908 | 1.17 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr1_+_199196059 | 1.17 |
ENSRNOT00000090428
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr19_-_25220010 | 1.16 |
ENSRNOT00000008786
|
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr1_+_192025710 | 1.14 |
ENSRNOT00000077457
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr5_-_60559329 | 1.11 |
ENSRNOT00000017046
|
Zbtb5
|
zinc finger and BTB domain containing 5 |
| chr1_-_167347662 | 1.07 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr5_+_2043583 | 1.06 |
ENSRNOT00000082813
|
Eloc
|
elongin C |
| chr3_-_157099306 | 1.04 |
ENSRNOT00000022861
|
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr6_+_28235695 | 1.03 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr1_+_154377247 | 1.03 |
ENSRNOT00000092945
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr8_+_75625174 | 1.02 |
ENSRNOT00000013742
|
Ice2
|
interactor of little elongation complex ELL subunit 2 |
| chr6_-_99783047 | 1.02 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr2_+_205553163 | 1.02 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr1_+_222311253 | 1.00 |
ENSRNOT00000028749
|
Macrod1
|
MACRO domain containing 1 |
| chr10_+_86711240 | 1.00 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chr13_-_89661150 | 0.99 |
ENSRNOT00000058390
|
Usp21
|
ubiquitin specific peptidase 21 |
| chr10_-_85124644 | 0.98 |
ENSRNOT00000012376
|
Kpnb1
|
karyopherin subunit beta 1 |
| chr1_-_73753128 | 0.97 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr2_+_151314818 | 0.96 |
ENSRNOT00000019305
|
P2ry1
|
purinergic receptor P2Y1 |
| chr13_-_110077946 | 0.96 |
ENSRNOT00000000078
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
| chrX_+_124894466 | 0.94 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr4_-_16130563 | 0.93 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr10_+_46906115 | 0.90 |
ENSRNOT00000034006
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr7_-_142132173 | 0.89 |
ENSRNOT00000026613
|
Csrnp2
|
cysteine and serine rich nuclear protein 2 |
| chr10_+_84214358 | 0.88 |
ENSRNOT00000011416
|
Hoxb1
|
homeo box B1 |
| chr10_+_56605140 | 0.87 |
ENSRNOT00000024079
|
Phf23
|
PHD finger protein 23 |
| chrX_+_27015884 | 0.85 |
ENSRNOT00000065814
|
Msl3
|
male-specific lethal 3 homolog (Drosophila) |
| chr9_-_82673898 | 0.84 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr13_-_94355219 | 0.83 |
ENSRNOT00000005332
|
Pld5
|
phospholipase D family, member 5 |
| chr1_-_167347490 | 0.83 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr2_-_188718704 | 0.82 |
ENSRNOT00000028010
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr10_+_110893457 | 0.82 |
ENSRNOT00000088495
|
Metrnl
|
meteorin-like, glial cell differentiation regulator |
| chr18_+_65814026 | 0.79 |
ENSRNOT00000016112
|
Mbd2
|
methyl-CpG binding domain protein 2 |
| chr3_+_110982553 | 0.78 |
ENSRNOT00000030754
|
LOC691418
|
hypothetical protein LOC691418 |
| chr10_+_82032656 | 0.73 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr7_-_119071712 | 0.71 |
ENSRNOT00000037611
|
Myh9l1
|
myosin heavy chain 9-like 1 |
| chr19_-_37667987 | 0.70 |
ENSRNOT00000059577
|
Acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chrX_-_33665821 | 0.70 |
ENSRNOT00000066676
|
Rbbp7
|
RB binding protein 7, chromatin remodeling factor |
| chr10_-_110182291 | 0.69 |
ENSRNOT00000015178
ENSRNOT00000054936 |
Csnk1d
|
casein kinase 1, delta |
| chr4_-_16130848 | 0.64 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr3_-_33075685 | 0.63 |
ENSRNOT00000006937
|
Orc4
|
origin recognition complex, subunit 4 |
| chr17_+_36335804 | 0.61 |
ENSRNOT00000091334
|
E2f3
|
E2F transcription factor 3 |
| chr16_+_20740826 | 0.60 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr2_-_188528127 | 0.59 |
ENSRNOT00000064715
ENSRNOT00000086683 |
Mtx1
|
Metaxin 1 |
| chr10_-_6870011 | 0.54 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr4_+_145017608 | 0.54 |
ENSRNOT00000066723
|
Setd5
|
SET domain containing 5 |
| chr10_+_88227360 | 0.53 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr13_-_48400632 | 0.53 |
ENSRNOT00000074204
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr18_-_37776453 | 0.52 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr17_-_77340934 | 0.51 |
ENSRNOT00000024697
|
Sephs1
|
selenophosphate synthetase 1 |
| chr1_-_88111293 | 0.48 |
ENSRNOT00000077195
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr1_-_218920094 | 0.47 |
ENSRNOT00000022213
|
Lrp5
|
LDL receptor related protein 5 |
| chr6_+_91532467 | 0.46 |
ENSRNOT00000082500
ENSRNOT00000064668 |
Klhdc1
|
kelch domain containing 1 |
| chr3_+_12262822 | 0.45 |
ENSRNOT00000022585
|
Angptl2
|
angiopoietin-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp219_Zfp740
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 2.0 | 6.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 1.8 | 7.2 | GO:0010424 | DNA methylation on cytosine within a CG sequence(GO:0010424) |
| 1.7 | 5.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 1.6 | 4.9 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 1.5 | 4.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.2 | 8.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.2 | 3.6 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.2 | 4.7 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 1.1 | 3.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.1 | 3.3 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 1.0 | 5.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 1.0 | 5.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.0 | 7.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 1.0 | 1.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.0 | 3.9 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.9 | 3.7 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.9 | 2.7 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.9 | 3.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.9 | 7.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.8 | 5.7 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.8 | 2.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.7 | 8.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.7 | 3.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.7 | 4.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.7 | 5.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.7 | 4.3 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.7 | 0.7 | GO:0070370 | cellular heat acclimation(GO:0070370) |
| 0.7 | 4.2 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.7 | 3.9 | GO:0035984 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.6 | 3.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.6 | 2.3 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.6 | 4.6 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.6 | 2.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.6 | 4.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.5 | 3.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 3.8 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.5 | 1.6 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.5 | 3.2 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.5 | 3.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 2.0 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.5 | 6.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 15.9 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.5 | 4.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.5 | 5.8 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.5 | 2.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.4 | 10.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.4 | 8.3 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.4 | 5.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.4 | 3.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 4.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 1.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.4 | 1.5 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 | 2.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.3 | 1.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.3 | 3.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 1.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 | 4.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.3 | 2.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 1.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 0.9 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 2.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.3 | 3.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 3.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 1.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.3 | 2.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 5.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.2 | 1.0 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 4.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 2.6 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.2 | 0.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 1.2 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.2 | 0.9 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 2.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 4.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 1.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 1.0 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.2 | 2.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 0.6 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 | 1.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 7.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 1.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 2.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 1.0 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 2.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 0.5 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.1 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 7.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 4.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 2.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 2.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.5 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 3.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.8 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 | 0.5 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 5.4 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 7.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.7 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 4.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.7 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 0.6 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 1.8 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 2.5 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.1 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 1.7 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 4.0 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 2.9 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 1.3 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.0 | 0.4 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.3 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 6.1 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 1.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.3 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 1.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.4 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.0 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.0 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.7 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.3 | GO:2001234 | negative regulation of apoptotic signaling pathway(GO:2001234) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.0 | 3.9 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.9 | 6.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.8 | 5.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.8 | 10.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.8 | 15.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.7 | 2.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.7 | 2.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.6 | 4.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.6 | 3.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 8.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.6 | 1.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 1.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.5 | 10.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 1.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 8.4 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.4 | 4.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 3.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 2.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 7.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 2.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 1.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.3 | 7.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 2.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 1.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.3 | 11.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 2.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.2 | 4.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 4.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 1.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 9.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.0 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 4.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 10.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 3.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 5.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 2.5 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 5.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 3.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 18.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 4.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 14.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 11.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.5 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0061700 | TORC2 complex(GO:0031932) GATOR2 complex(GO:0061700) |
| 0.0 | 1.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 4.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.2 | 4.9 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.8 | 5.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.6 | 5.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.6 | 6.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.6 | 5.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.5 | 8.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 1.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.5 | 3.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.5 | 1.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.5 | 2.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.5 | 2.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.5 | 2.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 3.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.5 | 4.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 3.7 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 5.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 8.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 7.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.4 | 8.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.4 | 1.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.4 | 3.9 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.3 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 4.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 11.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 2.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 4.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 5.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 3.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.3 | 3.0 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.3 | 2.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 3.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 1.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 14.1 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 1.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 3.9 | GO:1990381 | misfolded protein binding(GO:0051787) ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 0.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 4.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 4.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 3.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 5.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 4.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.8 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 5.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 10.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 4.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 2.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 2.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.9 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.8 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 3.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 17.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 3.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 7.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 6.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 3.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 9.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 3.4 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.1 | 11.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 4.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.0 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 6.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 7.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 3.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 7.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 6.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 3.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 2.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 2.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 7.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 7.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 4.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 4.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 17.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 4.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 5.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 1.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 4.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 4.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 3.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 2.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 10.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 18.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 3.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |