Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
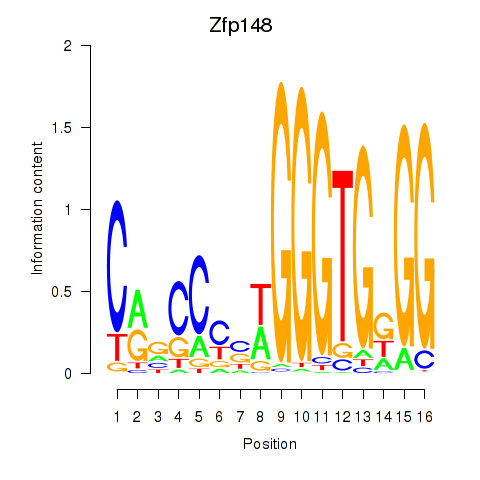
Results for Zfp148
Z-value: 1.15
Transcription factors associated with Zfp148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp148
|
ENSRNOG00000001789 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp148 | rn6_v1_chr11_-_70618347_70618347 | 0.46 | 1.7e-18 | Click! |
Activity profile of Zfp148 motif
Sorted Z-values of Zfp148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_113257688 | 38.74 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr2_-_29768750 | 37.25 |
ENSRNOT00000023460
|
Map1b
|
microtubule-associated protein 1B |
| chr14_-_18839420 | 37.16 |
ENSRNOT00000034090
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr7_+_133400485 | 36.71 |
ENSRNOT00000006219
|
Cntn1
|
contactin 1 |
| chr1_-_81881549 | 32.09 |
ENSRNOT00000027497
|
Atp1a3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr16_+_10417185 | 30.56 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr16_-_20686317 | 30.52 |
ENSRNOT00000060097
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr10_+_91710495 | 29.64 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr16_+_3851270 | 27.26 |
ENSRNOT00000014964
|
Plac9
|
placenta-specific 9 |
| chrX_+_39711201 | 26.90 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr1_-_89483988 | 25.06 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr14_+_84306466 | 24.86 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr10_-_90999506 | 22.75 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr4_+_56711049 | 22.53 |
ENSRNOT00000027237
|
Flnc
|
filamin C |
| chr4_+_21317695 | 22.39 |
ENSRNOT00000007572
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr1_-_215536980 | 21.10 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr2_+_54191538 | 20.75 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr4_+_56711275 | 20.67 |
ENSRNOT00000088149
|
Flnc
|
filamin C |
| chr1_-_7480825 | 20.67 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr8_+_59278262 | 20.29 |
ENSRNOT00000017053
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr4_-_14490446 | 19.61 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr5_-_109651730 | 19.39 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr5_+_147323240 | 18.89 |
ENSRNOT00000047152
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr5_-_141242131 | 18.62 |
ENSRNOT00000081482
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr9_+_118842787 | 18.42 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr10_-_61361250 | 17.83 |
ENSRNOT00000092203
|
Rap1gap2
|
RAP1 GTPase activating protein 2 |
| chr3_+_113818872 | 17.48 |
ENSRNOT00000044158
|
Casc4
|
cancer susceptibility candidate 4 |
| chr7_-_121232741 | 17.22 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr10_-_31258105 | 16.63 |
ENSRNOT00000008544
|
Nipal4
|
NIPA-like domain containing 4 |
| chr4_-_114853868 | 16.43 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr4_-_16130848 | 16.23 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr8_+_130283542 | 15.86 |
ENSRNOT00000071573
|
Vipr1
|
vasoactive intestinal peptide receptor 1 |
| chr6_+_1657331 | 15.86 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chrX_-_124464963 | 15.58 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr7_+_70364813 | 15.42 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr4_-_16130563 | 15.10 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr11_+_86903122 | 14.86 |
ENSRNOT00000048063
|
Zdhhc8
|
zinc finger, DHHC-type containing 8 |
| chr3_-_2689084 | 14.53 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr1_-_146556171 | 14.42 |
ENSRNOT00000017636
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr7_-_136853957 | 13.78 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr1_+_89491654 | 13.72 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr1_+_282265370 | 13.65 |
ENSRNOT00000015687
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr14_-_82287108 | 13.59 |
ENSRNOT00000023144
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr11_+_87722350 | 13.41 |
ENSRNOT00000000313
|
Scarf2
|
scavenger receptor class F, member 2 |
| chr10_+_59529785 | 13.41 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_-_239057732 | 13.39 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr6_+_137164535 | 13.39 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr14_-_36554580 | 13.27 |
ENSRNOT00000002869
|
Rasl11b
|
RAS-like family 11 member B |
| chr8_-_98738446 | 13.13 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr6_+_76675418 | 12.32 |
ENSRNOT00000076169
ENSRNOT00000010948 ENSRNOT00000082286 |
Brms1l
Brms1l
|
breast cancer metastasis-suppressor 1-like breast cancer metastasis-suppressor 1-like |
| chr2_+_188844073 | 11.79 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr8_+_26413233 | 11.20 |
ENSRNOT00000063967
ENSRNOT00000008838 |
Sept7
|
septin 7 |
| chr7_-_14189688 | 11.19 |
ENSRNOT00000037456
|
Notch3
|
notch 3 |
| chr8_-_59239954 | 11.18 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr13_-_72744295 | 11.11 |
ENSRNOT00000093366
ENSRNOT00000077157 |
Ier5
|
immediate early response 5 |
| chr7_-_70481796 | 11.02 |
ENSRNOT00000006815
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr14_-_18839595 | 10.94 |
ENSRNOT00000078746
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr1_-_188245628 | 10.89 |
ENSRNOT00000022800
|
Tmc7
|
transmembrane channel-like 7 |
| chr19_+_2393059 | 10.71 |
ENSRNOT00000018535
|
Cdh11
|
cadherin 11 |
| chr20_+_2194709 | 10.66 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr4_-_129515435 | 10.51 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr10_+_92289107 | 10.46 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr10_+_92288910 | 10.11 |
ENSRNOT00000006947
ENSRNOT00000045127 |
Mapt
|
microtubule-associated protein tau |
| chr1_+_226091774 | 9.66 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr5_+_103479767 | 9.49 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chrX_-_142131545 | 9.42 |
ENSRNOT00000077402
|
Fgf13
|
fibroblast growth factor 13 |
| chrX_+_15035569 | 9.38 |
ENSRNOT00000006491
ENSRNOT00000078392 |
Porcn
|
porcupine homolog (Drosophila) |
| chr14_+_114152472 | 9.32 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr1_-_215536770 | 9.25 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr2_-_144467912 | 9.21 |
ENSRNOT00000040002
|
Ccna1
|
cyclin A1 |
| chr4_-_113988246 | 9.20 |
ENSRNOT00000013128
|
LOC103692166
|
WD repeat-containing protein 54 |
| chr8_+_118013612 | 8.93 |
ENSRNOT00000056166
|
Map4
|
microtubule-associated protein 4 |
| chr7_-_120636563 | 8.83 |
ENSRNOT00000091317
ENSRNOT00000031117 |
Tmem184b
|
transmembrane protein 184B |
| chr4_+_85662892 | 8.82 |
ENSRNOT00000016175
ENSRNOT00000043851 ENSRNOT00000035722 ENSRNOT00000046192 |
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor type 1 |
| chr5_-_139227196 | 8.74 |
ENSRNOT00000050941
|
Foxo6
|
forkhead box O6 |
| chr6_-_137733026 | 8.53 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr5_+_59128315 | 8.43 |
ENSRNOT00000021802
|
Npr2
|
natriuretic peptide receptor 2 |
| chr10_-_39405311 | 8.25 |
ENSRNOT00000074616
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr10_+_108340240 | 8.10 |
ENSRNOT00000077535
|
Ccdc40
|
coiled-coil domain containing 40 |
| chr8_-_116391158 | 8.07 |
ENSRNOT00000078720
ENSRNOT00000022550 |
Gnai2
|
G protein subunit alpha i2 |
| chr1_+_157920786 | 7.99 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr17_-_55709740 | 7.95 |
ENSRNOT00000033359
|
RGD1562037
|
similar to OTTHUMP00000046255 |
| chr3_-_112789282 | 7.86 |
ENSRNOT00000090680
ENSRNOT00000066181 |
Ttbk2
|
tau tubulin kinase 2 |
| chr7_-_143738237 | 7.84 |
ENSRNOT00000055320
|
Spryd3
|
SPRY domain containing 3 |
| chr19_-_58399816 | 7.75 |
ENSRNOT00000026843
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr7_-_141424225 | 7.67 |
ENSRNOT00000080109
ENSRNOT00000084292 |
Cers5
|
ceramide synthase 5 |
| chr7_-_116607408 | 7.65 |
ENSRNOT00000076009
ENSRNOT00000056554 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr12_+_16170162 | 7.39 |
ENSRNOT00000001686
|
Grifin
|
galectin-related inter-fiber protein |
| chr3_-_63535991 | 7.25 |
ENSRNOT00000015722
|
Fkbp7
|
FK506 binding protein 7 |
| chrX_+_119030419 | 7.22 |
ENSRNOT00000060168
|
Pls3
|
plastin 3 |
| chr9_-_46401911 | 7.17 |
ENSRNOT00000046557
|
Creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr1_+_220668544 | 6.78 |
ENSRNOT00000039842
|
Gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr7_+_130474508 | 6.68 |
ENSRNOT00000085191
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_-_103256823 | 6.66 |
ENSRNOT00000018860
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr8_-_125645898 | 6.50 |
ENSRNOT00000036672
|
Rbms3
|
RNA binding motif, single stranded interacting protein 3 |
| chr13_+_52588917 | 6.42 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr14_+_36687134 | 6.13 |
ENSRNOT00000002879
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr8_-_94563760 | 6.03 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chr14_-_80732010 | 5.98 |
ENSRNOT00000012322
|
Adra2c
|
adrenoceptor alpha 2C |
| chr2_+_211078334 | 5.95 |
ENSRNOT00000049127
|
Sort1
|
sortilin 1 |
| chr4_-_100883275 | 5.93 |
ENSRNOT00000022846
|
LOC100364435
|
thymosin, beta 10-like |
| chr7_+_41114697 | 5.81 |
ENSRNOT00000041354
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr8_+_117246376 | 5.77 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr9_+_82674202 | 5.56 |
ENSRNOT00000027208
|
Tmem198
|
transmembrane protein 198 |
| chr5_-_151072766 | 5.54 |
ENSRNOT00000078048
ENSRNOT00000017227 ENSRNOT00000085573 |
Stx12
|
syntaxin 12 |
| chr5_-_166726794 | 5.54 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr7_+_130474279 | 5.46 |
ENSRNOT00000092388
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr13_+_99173484 | 5.45 |
ENSRNOT00000080574
ENSRNOT00000088654 |
Lefty2
|
Left-right determination factor 2 |
| chr4_-_100883038 | 5.45 |
ENSRNOT00000041880
|
LOC100364435
|
thymosin, beta 10-like |
| chr5_+_36076565 | 5.39 |
ENSRNOT00000013599
|
Faxc
|
failed axon connections homolog |
| chr12_+_48481750 | 5.35 |
ENSRNOT00000000886
|
Coro1c
|
coronin 1C |
| chr7_-_117680004 | 5.33 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr9_+_17340341 | 5.24 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr19_-_55510460 | 5.22 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr9_-_45292047 | 5.15 |
ENSRNOT00000083256
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr4_-_118595580 | 5.07 |
ENSRNOT00000024436
|
Anxa4
|
annexin A4 |
| chr19_+_40855433 | 5.06 |
ENSRNOT00000023823
|
Il34
|
interleukin 34 |
| chr3_-_177201525 | 5.05 |
ENSRNOT00000022451
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr13_-_48848864 | 5.04 |
ENSRNOT00000077857
ENSRNOT00000068003 |
Mfsd4
|
major facilitator superfamily domain containing 4 |
| chr9_+_82647071 | 5.02 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr10_+_10644572 | 4.99 |
ENSRNOT00000004026
|
Ppl
|
periplakin |
| chr4_-_83137527 | 4.89 |
ENSRNOT00000039580
|
Jazf1
|
JAZF zinc finger 1 |
| chr7_-_142132173 | 4.85 |
ENSRNOT00000026613
|
Csrnp2
|
cysteine and serine rich nuclear protein 2 |
| chr9_-_82673898 | 4.81 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr3_-_81886545 | 4.79 |
ENSRNOT00000057007
|
LOC102547078
|
myb-related transcription factor, partner of profilin-like |
| chrX_+_24891156 | 4.75 |
ENSRNOT00000071173
|
Wwc3
|
WWC family member 3 |
| chr1_+_87790104 | 4.75 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr1_+_202432366 | 4.70 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chrX_+_156463953 | 4.41 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr2_+_196334626 | 4.23 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr6_-_27190126 | 4.15 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr12_+_47074200 | 4.12 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr2_+_198823366 | 4.00 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr2_+_62150251 | 3.99 |
ENSRNOT00000016196
|
Zfr
|
zinc finger RNA binding protein |
| chr13_-_48284990 | 3.98 |
ENSRNOT00000086928
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr12_-_41448668 | 3.96 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr10_-_74679858 | 3.92 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr4_+_114854458 | 3.83 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr2_-_210413985 | 3.48 |
ENSRNOT00000025036
|
Strip1
|
striatin interacting protein 1 |
| chr5_+_59063531 | 3.46 |
ENSRNOT00000085311
ENSRNOT00000065909 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr7_-_139318455 | 3.42 |
ENSRNOT00000092029
|
Hdac7
|
histone deacetylase 7 |
| chr1_-_222734184 | 3.35 |
ENSRNOT00000049812
ENSRNOT00000028785 |
Rtn3
|
reticulon 3 |
| chr17_+_9837402 | 3.35 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr6_-_38228379 | 3.32 |
ENSRNOT00000084924
|
Mycn
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr1_-_215836641 | 3.30 |
ENSRNOT00000080246
|
Igf2
|
insulin-like growth factor 2 |
| chr1_-_142615673 | 3.29 |
ENSRNOT00000018021
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr19_+_14352411 | 3.16 |
ENSRNOT00000067519
|
Hmgxb4
|
HMG-box containing 4 |
| chr3_+_11629556 | 3.13 |
ENSRNOT00000074401
|
St6galnac6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr1_+_201055644 | 3.10 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_23581052 | 3.07 |
ENSRNOT00000006190
|
Ppp1cb
|
protein phosphatase 1 catalytic subunit beta |
| chr5_-_58163584 | 3.03 |
ENSRNOT00000060594
|
Ccl27
|
C-C motif chemokine ligand 27 |
| chr11_+_38035611 | 3.01 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr10_+_56453877 | 2.98 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr10_-_6870011 | 2.97 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr13_+_34365147 | 2.96 |
ENSRNOT00000093066
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr12_-_18540166 | 2.94 |
ENSRNOT00000001792
|
Il3ra
|
interleukin 3 receptor subunit alpha |
| chr1_+_266105124 | 2.90 |
ENSRNOT00000043925
ENSRNOT00000088243 |
Mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr10_-_13446135 | 2.71 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr20_-_3438389 | 2.63 |
ENSRNOT00000001098
ENSRNOT00000090039 |
Flot1
|
flotillin 1 |
| chr6_-_86822094 | 2.43 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr20_-_8574082 | 2.39 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr6_+_111049559 | 2.37 |
ENSRNOT00000015571
|
Tmem63c
|
transmembrane protein 63c |
| chr7_+_140716113 | 2.35 |
ENSRNOT00000033450
|
Tuba1c
|
tubulin, alpha 1C |
| chr3_-_112061796 | 2.30 |
ENSRNOT00000056258
|
Pla2g4d
|
phospholipase A2 group IVD |
| chr15_+_33555640 | 2.20 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr11_+_38035450 | 2.04 |
ENSRNOT00000083067
|
Mx2
|
MX dynamin like GTPase 2 |
| chr19_+_10119253 | 2.01 |
ENSRNOT00000017971
|
Zfp319
|
zinc finger protein 319 |
| chr1_-_102849430 | 1.99 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr1_-_82108083 | 1.92 |
ENSRNOT00000027677
ENSRNOT00000084530 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr20_-_5618254 | 1.92 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr6_+_28235695 | 1.90 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr17_-_14091766 | 1.88 |
ENSRNOT00000075373
|
AABR07027088.1
|
|
| chr1_+_102849889 | 1.78 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr5_-_154223536 | 1.75 |
ENSRNOT00000012258
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr10_-_85574889 | 1.39 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr12_+_2069959 | 1.33 |
ENSRNOT00000001298
|
Pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr7_+_35069814 | 1.31 |
ENSRNOT00000089228
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr1_+_216237697 | 1.27 |
ENSRNOT00000027760
|
Cd81
|
Cd81 molecule |
| chr10_+_93811350 | 1.26 |
ENSRNOT00000077280
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr14_-_85350786 | 1.17 |
ENSRNOT00000012634
|
LOC100912481
|
RNA-binding protein EWS-like |
| chr10_+_94280703 | 1.13 |
ENSRNOT00000088656
|
Map3k3
|
mitogen activated protein kinase kinase kinase 3 |
| chr2_+_62150493 | 1.13 |
ENSRNOT00000080241
|
Zfr
|
zinc finger RNA binding protein |
| chr8_-_36467627 | 1.06 |
ENSRNOT00000082346
|
Fam118b
|
family with sequence similarity 118, member B |
| chr16_+_74292438 | 1.00 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr4_+_100407658 | 0.96 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr1_+_46378807 | 0.93 |
ENSRNOT00000043521
|
Zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr8_-_23084879 | 0.92 |
ENSRNOT00000018874
|
Zfp653
|
zinc finger protein 653 |
| chr1_+_101012822 | 0.89 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr7_-_144880092 | 0.72 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr4_+_24612205 | 0.67 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr14_-_84937725 | 0.67 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_+_282134981 | 0.64 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr2_-_232245319 | 0.57 |
ENSRNOT00000014510
|
LOC691931
|
hypothetical protein LOC691931 |
| chrX_+_71155601 | 0.56 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr7_+_35472102 | 0.52 |
ENSRNOT00000010316
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr19_+_25969255 | 0.46 |
ENSRNOT00000004370
|
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr6_-_146195819 | 0.44 |
ENSRNOT00000007625
|
Sp4
|
Sp4 transcription factor |
| chr4_-_152380184 | 0.39 |
ENSRNOT00000091473
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chrX_+_69730242 | 0.37 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr8_-_116635851 | 0.35 |
ENSRNOT00000024939
|
Rbm6
|
RNA binding motif protein 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp148
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 30.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 5.7 | 17.2 | GO:0072254 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 5.3 | 15.9 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 5.3 | 31.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 5.2 | 31.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 4.9 | 19.6 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 4.4 | 30.6 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 3.8 | 22.7 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 3.7 | 11.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 3.6 | 14.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 3.6 | 10.7 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 3.2 | 32.1 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 3.0 | 12.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 3.0 | 8.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 2.8 | 35.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 2.7 | 13.6 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 2.4 | 18.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 2.3 | 18.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 2.3 | 18.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 2.2 | 11.2 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 2.1 | 24.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 2.0 | 22.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 2.0 | 20.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.8 | 5.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.7 | 5.2 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 1.7 | 13.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.7 | 8.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 1.6 | 8.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 1.5 | 6.0 | GO:0071883 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 1.4 | 11.2 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 1.3 | 8.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 1.3 | 51.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.3 | 48.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 1.3 | 4.0 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 1.2 | 8.5 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.2 | 6.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.2 | 16.6 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 1.2 | 9.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.2 | 8.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.2 | 3.5 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 1.1 | 6.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 1.1 | 3.3 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 1.0 | 5.8 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 1.0 | 1.9 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.9 | 4.4 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.9 | 6.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.9 | 13.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.8 | 5.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.8 | 4.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.8 | 10.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.8 | 5.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.7 | 5.2 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.7 | 36.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.7 | 10.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.7 | 5.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.7 | 3.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.7 | 8.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.7 | 3.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.7 | 3.3 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.7 | 2.6 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.7 | 5.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.6 | 1.3 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) |
| 0.6 | 1.9 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.6 | 43.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.6 | 5.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.6 | 7.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.5 | 13.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.5 | 6.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.5 | 13.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.5 | 15.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.5 | 2.9 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.5 | 1.9 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.5 | 2.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.5 | 3.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 3.0 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.4 | 1.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.3 | 3.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.3 | 4.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 4.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.3 | 13.1 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.3 | 7.6 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 3.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 3.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 13.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.2 | 3.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 13.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 5.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 9.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 0.6 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 8.7 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.2 | 10.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.2 | 0.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.2 | 9.4 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.2 | 3.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 4.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 4.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 6.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 11.8 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 5.5 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 4.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.6 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.1 | 7.7 | GO:0046467 | membrane lipid biosynthetic process(GO:0046467) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 2.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 21.0 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.7 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 1.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 4.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.0 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 1.0 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 4.2 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 30.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 5.7 | 22.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 5.3 | 32.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 5.1 | 20.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.6 | 31.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 2.2 | 11.2 | GO:0005940 | septin ring(GO:0005940) |
| 2.0 | 6.0 | GO:0098833 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 1.8 | 5.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.3 | 26.2 | GO:0043196 | varicosity(GO:0043196) |
| 1.2 | 22.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.0 | 43.2 | GO:0043034 | costamere(GO:0043034) |
| 0.7 | 3.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.7 | 6.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.7 | 8.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 4.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.4 | 5.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 38.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.4 | 11.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 3.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.4 | 30.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.4 | 13.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 9.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.4 | 12.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.4 | 37.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 7.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 3.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 9.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 4.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 6.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 73.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.2 | 7.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 12.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 6.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 6.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 1.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 2.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 11.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 5.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 4.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 15.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 11.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 11.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 13.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.9 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 4.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 8.1 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 5.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 14.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 9.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 2.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 15.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 5.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 15.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 5.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 12.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 6.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 9.0 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 30.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 7.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 15.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 21.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 4.3 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 31.3 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 8.2 | 24.7 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 7.5 | 22.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 5.1 | 20.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 5.1 | 30.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 3.4 | 48.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.8 | 8.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 2.4 | 30.6 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 2.3 | 32.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 2.3 | 13.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 2.1 | 10.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 2.0 | 6.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.8 | 23.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.7 | 17.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.7 | 13.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.7 | 6.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.7 | 5.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.7 | 11.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.6 | 37.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 1.5 | 6.0 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.2 | 14.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 1.1 | 43.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.1 | 11.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.1 | 16.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.0 | 7.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.9 | 19.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.8 | 4.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.8 | 34.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 9.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 4.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.7 | 20.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.7 | 6.0 | GO:0098748 | clathrin heavy chain binding(GO:0032050) clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.7 | 15.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.6 | 3.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 4.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.6 | 5.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.6 | 3.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 7.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 15.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.5 | 13.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.5 | 15.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.5 | 38.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 14.5 | GO:0005501 | retinoid binding(GO:0005501) |
| 0.4 | 1.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 8.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 42.8 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.3 | 9.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 4.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 12.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 12.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 12.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 6.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 59.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 6.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 20.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.2 | 18.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 1.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 4.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 3.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 10.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 5.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 5.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 8.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 42.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 3.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 3.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 5.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 18.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 7.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 2.6 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 6.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 23.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 7.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 3.7 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 20.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.5 | 30.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 1.2 | 52.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.9 | 39.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 55.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.4 | 5.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.4 | 66.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 13.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 4.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.3 | 12.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 13.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 9.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 78.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 2.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 3.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 4.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 15.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 1.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 11.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 14.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 5.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 43.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.4 | 36.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 2.0 | 8.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 1.6 | 19.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.3 | 13.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 1.0 | 17.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 1.0 | 13.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 1.0 | 51.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 1.0 | 22.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.9 | 45.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.7 | 20.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 9.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.6 | 24.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.4 | 5.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 9.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 10.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 4.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 14.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.3 | 5.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 5.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 3.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 5.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 4.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 16.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.2 | 7.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 5.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 4.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.3 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 1.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 3.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 6.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 11.8 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 5.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 1.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |