Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
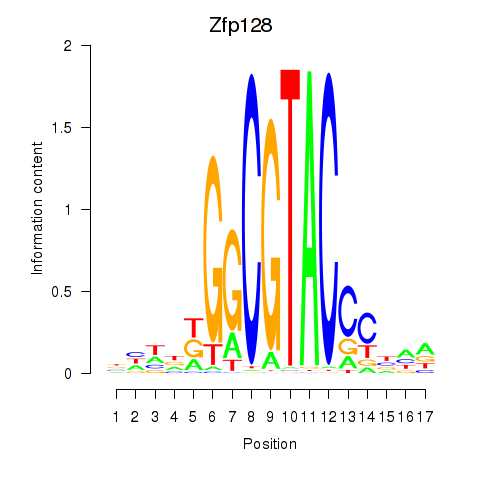
Results for Zfp128
Z-value: 0.41
Transcription factors associated with Zfp128
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp128
|
ENSRNOG00000031113 | zinc finger protein 128 |
|
Zfp128
|
ENSRNOG00000031459 | zinc finger protein 128 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp128 | rn6_v1_chr1_-_65701867_65701867 | -0.18 | 1.1e-03 | Click! |
Activity profile of Zfp128 motif
Sorted Z-values of Zfp128 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_23256158 | 13.20 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr10_+_11240138 | 9.29 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr13_+_75111778 | 8.88 |
ENSRNOT00000006924
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr6_-_23316962 | 8.48 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_-_78985990 | 8.31 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr8_-_130550388 | 8.12 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr1_+_101427195 | 6.93 |
ENSRNOT00000028271
|
Gys1
|
glycogen synthase 1 |
| chrX_+_156999826 | 6.11 |
ENSRNOT00000081819
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD), gamma |
| chr10_+_102136283 | 5.97 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr5_+_151692108 | 4.82 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr1_-_170471076 | 3.81 |
ENSRNOT00000025159
|
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr2_+_210685197 | 3.75 |
ENSRNOT00000072342
|
NEWGENE_620381
|
glutathione S-transferase mu 3 |
| chr1_+_170471238 | 3.68 |
ENSRNOT00000076961
ENSRNOT00000075597 ENSRNOT00000076631 ENSRNOT00000076783 |
Timm10b
Dnhd1
|
translocase of inner mitochondrial membrane 10B dynein heavy chain domain 1 |
| chr8_-_59629133 | 3.14 |
ENSRNOT00000019458
|
Chrnb4
|
cholinergic receptor nicotinic beta 4 subunit |
| chr6_-_128989812 | 3.03 |
ENSRNOT00000085943
|
LOC100909439
|
ankyrin repeat and SOCS box protein 2-like |
| chr2_-_210839295 | 2.80 |
ENSRNOT00000025994
|
Gstm4
|
glutathione S-transferase mu 4 |
| chr8_-_62215293 | 2.38 |
ENSRNOT00000080448
|
NEWGENE_1306267
|
phosphopantothenoylcysteine decarboxylase |
| chr10_+_110631494 | 2.29 |
ENSRNOT00000054915
|
Fn3k
|
fructosamine 3 kinase |
| chr5_-_57168610 | 2.00 |
ENSRNOT00000090499
|
B4galt1
|
beta-1,4-galactosyltransferase 1 |
| chr8_+_111107358 | 1.88 |
ENSRNOT00000011203
|
Anapc13
|
anaphase promoting complex subunit 13 |
| chr20_+_32717564 | 1.55 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr14_+_2892753 | 1.44 |
ENSRNOT00000061630
|
Evi5
|
ecotropic viral integration site 5 |
| chr5_-_147867583 | 1.38 |
ENSRNOT00000000139
|
Kpna6
|
karyopherin subunit alpha 6 |
| chr4_-_150616895 | 1.19 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr3_-_177089199 | 0.98 |
ENSRNOT00000020873
|
Zfp512b
|
zinc finger protein 512B |
| chr7_+_117390285 | 0.95 |
ENSRNOT00000016464
|
Exosc4
|
exosome component 4 |
| chr1_-_102970950 | 0.92 |
ENSRNOT00000018194
|
Tsg101
|
tumor susceptibility 101 |
| chr8_-_21384131 | 0.83 |
ENSRNOT00000071010
|
Zfp560
|
zinc finger protein 560 |
| chr8_-_64154396 | 0.76 |
ENSRNOT00000031262
|
Bbs4
|
Bardet-Biedl syndrome 4 |
| chr7_-_19088972 | 0.74 |
ENSRNOT00000049266
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr10_-_71383378 | 0.70 |
ENSRNOT00000077025
|
Dusp14
|
dual specificity phosphatase 14 |
| chr12_-_51341663 | 0.66 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr18_+_41022552 | 0.65 |
ENSRNOT00000005260
|
Commd10
|
COMM domain containing 10 |
| chr1_+_162320730 | 0.64 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr1_-_170186462 | 0.61 |
ENSRNOT00000035798
|
Olr202
|
olfactory receptor 202 |
| chr10_-_71383602 | 0.55 |
ENSRNOT00000083300
|
Dusp14
|
dual specificity phosphatase 14 |
| chr20_-_31598118 | 0.52 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr6_+_18877907 | 0.50 |
ENSRNOT00000084222
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr20_-_13581164 | 0.45 |
ENSRNOT00000033298
|
Zfp280b
|
zinc finger protein 280B |
| chr7_-_22943159 | 0.40 |
ENSRNOT00000047547
|
Vom2r57
|
vomeronasal 2 receptor, 57 |
| chr3_-_8690305 | 0.39 |
ENSRNOT00000035002
|
Zer1
|
zyg-11 related, cell cycle regulator |
| chr4_+_155532109 | 0.37 |
ENSRNOT00000077245
|
Nanog
|
Nanog homeobox |
| chr4_+_155531906 | 0.31 |
ENSRNOT00000060937
|
Nanog
|
Nanog homeobox |
| chr5_+_103251986 | 0.30 |
ENSRNOT00000008757
|
Cntln
|
centlein |
| chr1_-_167971151 | 0.30 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chrX_+_73475049 | 0.28 |
ENSRNOT00000050601
|
Zfp36l3
|
zinc finger protein 36, C3H type-like 3 |
| chrX_-_156999650 | 0.22 |
ENSRNOT00000083557
|
Ssr4
|
signal sequence receptor subunit 4 |
| chr1_-_174350196 | 0.15 |
ENSRNOT00000055158
|
LOC499240
|
similar to predicted gene ICRFP703B1614Q5.5 |
| chr8_+_23014956 | 0.13 |
ENSRNOT00000018009
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr4_+_99398494 | 0.05 |
ENSRNOT00000064374
|
Rnf103
|
ring finger protein 103 |
| chr8_+_13305152 | 0.03 |
ENSRNOT00000012940
|
Mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr11_-_45124423 | 0.03 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chrX_-_57058526 | 0.01 |
ENSRNOT00000091191
|
AABR07038624.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp128
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.2 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 2.3 | 9.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.0 | 3.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 1.0 | 6.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.0 | 6.0 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.9 | 2.8 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.8 | 8.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.5 | 2.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 0.9 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 3.8 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 0.8 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 8.9 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 1.0 | GO:0071051 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 6.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.7 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.3 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 3.2 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0045242 | mitochondrial isocitrate dehydrogenase complex (NAD+)(GO:0005962) isocitrate dehydrogenase complex (NAD+)(GO:0045242) |
| 0.7 | 3.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 2.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 3.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 6.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 8.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 8.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 8.3 | GO:0019862 | IgA binding(GO:0019862) |
| 1.7 | 13.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.5 | 6.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.5 | 8.9 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.0 | 6.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.0 | 6.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.7 | 2.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.3 | 8.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 3.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 2.8 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.7 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 3.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 3.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 9.3 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 8.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 8.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 13.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 6.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 3.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 6.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 6.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.7 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |