Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
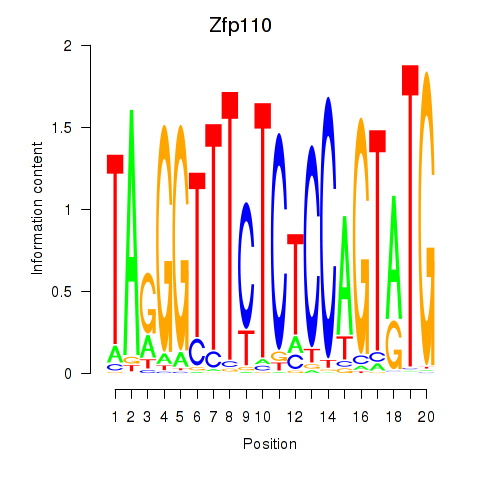
Results for Zfp110
Z-value: 1.38
Transcription factors associated with Zfp110
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp110
|
ENSRNOG00000031328 | zinc finger protein 110 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp110 | rn6_v1_chr1_-_65756528_65756528 | 0.26 | 1.8e-06 | Click! |
Activity profile of Zfp110 motif
Sorted Z-values of Zfp110 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_15821927 | 105.13 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr1_-_62316450 | 83.97 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr1_-_62558033 | 77.71 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr1_-_87777359 | 62.09 |
ENSRNOT00000074090
|
AABR07002870.1
|
|
| chr12_-_37185548 | 53.86 |
ENSRNOT00000001338
|
LOC102556092
|
uncharacterized LOC102556092 |
| chr1_-_88558696 | 52.58 |
ENSRNOT00000038642
|
AABR07002896.1
|
|
| chr7_-_15852930 | 52.10 |
ENSRNOT00000009270
|
LOC691422
|
similar to zinc finger protein 101 |
| chr14_+_6298872 | 46.76 |
ENSRNOT00000065071
|
Znf442
|
zinc finger protein 442 |
| chr1_+_88542035 | 42.07 |
ENSRNOT00000032973
|
Zfp420
|
zinc finger protein 420 |
| chr4_+_170347410 | 35.84 |
ENSRNOT00000040508
|
LOC500350
|
LRRGT00139 |
| chr1_-_87777668 | 35.78 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr1_+_84758233 | 31.02 |
ENSRNOT00000072734
|
AABR07002775.1
|
|
| chr5_-_164648328 | 30.10 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chr1_-_62191818 | 27.02 |
ENSRNOT00000033187
|
AABR07001926.1
|
|
| chr7_-_9976367 | 24.97 |
ENSRNOT00000044241
|
Zfp347
|
zinc finger protein 347 |
| chr7_+_15279611 | 23.69 |
ENSRNOT00000043826
|
Zfp952
|
zinc finger protein 952 |
| chr1_-_62114672 | 23.27 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr1_-_88558387 | 22.30 |
ENSRNOT00000074174
|
AABR07002896.1
|
|
| chr7_-_10588202 | 22.01 |
ENSRNOT00000086920
|
AABR07055826.1
|
|
| chr1_+_84652632 | 21.88 |
ENSRNOT00000041861
|
AABR07002774.2
|
|
| chrX_-_77295426 | 16.97 |
ENSRNOT00000090833
|
Taf9b
|
TATA-box binding protein associated factor 9b |
| chr1_+_61374379 | 15.76 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr8_+_131965134 | 15.00 |
ENSRNOT00000078308
|
Znf660
|
zinc finger protein 660 |
| chr4_+_110699557 | 13.76 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr20_+_1749716 | 12.65 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr20_+_1764794 | 12.11 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr2_+_230901126 | 12.07 |
ENSRNOT00000016026
ENSRNOT00000015564 ENSRNOT00000068198 |
Camk2d
|
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_282492912 | 11.97 |
ENSRNOT00000047883
|
AABR07007130.1
|
|
| chr1_+_88451958 | 11.74 |
ENSRNOT00000074301
|
LOC102549115
|
zinc finger protein 260-like |
| chr1_+_80973739 | 10.76 |
ENSRNOT00000026088
ENSRNOT00000081983 |
AC118165.1
|
|
| chr20_+_4369178 | 9.78 |
ENSRNOT00000088079
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr2_-_264864265 | 9.60 |
ENSRNOT00000044236
|
Srsf11
|
serine and arginine rich splicing factor 11 |
| chr15_-_48445588 | 8.99 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr18_+_79813759 | 8.86 |
ENSRNOT00000021768
|
Zfp516
|
zinc finger protein 516 |
| chrX_+_43535737 | 8.52 |
ENSRNOT00000087073
|
LOC102557137
|
E3 ubiquitin-protein ligase RNF168-like |
| chr16_-_21473808 | 8.34 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chr9_+_50892605 | 8.23 |
ENSRNOT00000033133
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr2_+_264864351 | 7.77 |
ENSRNOT00000015434
|
Lrrc40
|
leucine rich repeat containing 40 |
| chr2_+_22950018 | 7.30 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr5_-_113939127 | 6.66 |
ENSRNOT00000039554
|
Mysm1
|
myb-like, SWIRM and MPN domains 1 |
| chr5_+_153197459 | 6.47 |
ENSRNOT00000023187
|
Rhd
|
Rh blood group, D antigen |
| chr6_+_29977797 | 6.46 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr20_+_1781320 | 6.37 |
ENSRNOT00000040388
|
Olr1737
|
olfactory receptor 1737 |
| chr10_+_59533480 | 5.99 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr4_-_173640684 | 5.73 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chr1_+_67340809 | 5.70 |
ENSRNOT00000045266
|
LOC100362054
|
mCG114696-like |
| chr5_-_99033107 | 5.55 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr1_+_69841998 | 5.23 |
ENSRNOT00000072231
|
Zfp773-ps1
|
zinc finger protein 773, pseudogene 1 |
| chr1_-_61872975 | 4.77 |
ENSRNOT00000078809
|
AABR07001910.1
|
|
| chrX_-_62698830 | 4.64 |
ENSRNOT00000076359
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr4_+_49369296 | 4.58 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr14_+_41388563 | 4.18 |
ENSRNOT00000048329
|
LOC360933
|
similar to Ac1591 |
| chr8_+_64531594 | 4.14 |
ENSRNOT00000036798
|
Gramd2
|
GRAM domain containing 2 |
| chr19_-_36157924 | 3.63 |
ENSRNOT00000072022
|
AABR07043701.1
|
|
| chr18_-_76653056 | 3.53 |
ENSRNOT00000078496
|
Adnp2
|
ADNP homeobox 2 |
| chr17_+_36690249 | 3.26 |
ENSRNOT00000089476
ENSRNOT00000082140 |
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr1_+_150130084 | 3.21 |
ENSRNOT00000041243
|
Olr25
|
olfactory receptor 25 |
| chr12_-_2661565 | 2.80 |
ENSRNOT00000064859
|
Cd209f
|
CD209f antigen |
| chr10_-_57739109 | 2.34 |
ENSRNOT00000082171
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr13_+_99004942 | 2.13 |
ENSRNOT00000085390
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr10_+_29606748 | 1.78 |
ENSRNOT00000080720
|
AABR07029467.2
|
|
| chr16_-_83278119 | 1.62 |
ENSRNOT00000019454
|
Ing1
|
inhibitor of growth family, member 1 |
| chr12_-_11157583 | 1.58 |
ENSRNOT00000082535
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr4_+_166986350 | 1.46 |
ENSRNOT00000040828
|
Tas2r117
|
taste receptor, type 2, member 117 |
| chr1_+_60248695 | 1.36 |
ENSRNOT00000079558
|
Vom1r9
|
vomeronasal 1 receptor 9 |
| chr1_-_199061107 | 1.27 |
ENSRNOT00000085954
|
Zfp689
|
zinc finger protein 689 |
| chr16_-_19583386 | 1.25 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr7_-_18180705 | 1.05 |
ENSRNOT00000065954
|
Zfp494
|
zinc finger protein 494 |
| chr5_-_99566317 | 0.93 |
ENSRNOT00000051028
ENSRNOT00000089235 |
Mpdz
|
multiple PDZ domain crumbs cell polarity complex component |
| chr3_-_105663457 | 0.42 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chr7_-_120518653 | 0.41 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr17_-_18634258 | 0.30 |
ENSRNOT00000082307
|
LOC102554666
|
zinc finger protein 709-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp110
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 1.9 | 5.6 | GO:0097214 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 1.7 | 6.7 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.2 | 35.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 1.2 | 4.6 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.1 | 13.8 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.9 | 6.5 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.7 | 9.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.7 | 4.6 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.5 | 7.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.4 | 6.5 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.4 | 17.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.4 | 9.0 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.2 | 8.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 2.3 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 3.3 | GO:0032392 | DNA geometric change(GO:0032392) |
| 0.0 | 3.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 123.6 | GO:0006355 | regulation of transcription, DNA-templated(GO:0006355) |
| 0.0 | 9.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.1 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 4.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 13.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.4 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.9 | GO:0042552 | myelination(GO:0042552) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 35.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.9 | 5.6 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 1.3 | 17.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.7 | 24.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.6 | 4.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.5 | 2.3 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.4 | 13.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 7.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 9.2 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 17.0 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 1.8 | 35.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.5 | 7.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.4 | 5.6 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 1.3 | 12.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.8 | 9.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.7 | 4.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.6 | 6.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.6 | 13.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 6.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 6.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 3.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 4.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 8.9 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 118.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 2.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 6.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 2.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 5.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 13.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 5.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 4.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.0 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 6.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 9.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 4.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 48.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.2 | 5.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 9.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 9.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 4.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |