Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
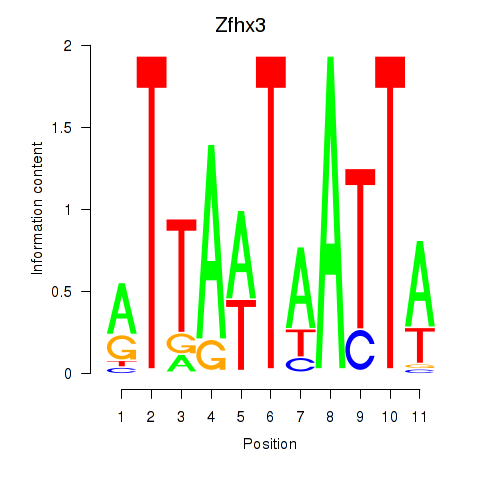
Results for Zfhx3
Z-value: 1.04
Transcription factors associated with Zfhx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfhx3
|
ENSRNOG00000014452 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfhx3 | rn6_v1_chr19_-_42920344_42920344 | 0.39 | 4.9e-13 | Click! |
Activity profile of Zfhx3 motif
Sorted Z-values of Zfhx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_14229067 | 57.91 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr13_+_56598957 | 55.95 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr6_+_80188943 | 55.66 |
ENSRNOT00000059335
|
Mia2
|
melanoma inhibitory activity 2 |
| chr6_+_127743971 | 50.30 |
ENSRNOT00000013045
|
Serpina4
|
serpin family A member 4 |
| chr17_-_43543172 | 47.48 |
ENSRNOT00000080684
ENSRNOT00000029626 ENSRNOT00000082719 |
Slc17a3
|
solute carrier family 17 member 3 |
| chr5_+_117698764 | 43.58 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr10_+_89285855 | 37.74 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr20_+_30690810 | 37.64 |
ENSRNOT00000000687
|
Pcbd1
|
pterin-4 alpha-carbinolamine dehydratase 1 |
| chr1_+_88955440 | 36.05 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr14_-_19132208 | 36.04 |
ENSRNOT00000060535
|
Afm
|
afamin |
| chr1_+_88955135 | 34.45 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr10_+_89286047 | 34.18 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr1_+_107262659 | 32.88 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr8_-_77398156 | 31.60 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr1_-_48563776 | 31.07 |
ENSRNOT00000023368
|
Plg
|
plasminogen |
| chr2_-_88763733 | 30.72 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_+_117422704 | 28.49 |
ENSRNOT00000085647
|
Slc12a1
|
solute carrier family 12 member 1 |
| chr9_+_95256627 | 26.60 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr4_-_176294997 | 25.64 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr12_-_29743705 | 25.52 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr14_-_80973456 | 25.43 |
ENSRNOT00000013257
|
Hgfac
|
HGF activator |
| chr2_+_235264219 | 25.28 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr10_-_103848035 | 23.71 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr2_-_243407608 | 23.54 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr13_+_91080341 | 23.05 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr1_+_261291870 | 21.93 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr16_+_7758996 | 19.55 |
ENSRNOT00000061063
|
Btd
|
biotinidase |
| chr1_-_126211439 | 17.69 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr2_-_88660449 | 13.20 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr2_-_166682325 | 13.11 |
ENSRNOT00000091198
ENSRNOT00000012422 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr1_-_266428239 | 12.71 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr13_-_86671515 | 12.20 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr5_+_8916633 | 11.55 |
ENSRNOT00000066395
|
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr13_+_99369126 | 11.38 |
ENSRNOT00000072695
|
LOC498300
|
similar to degenerative spermatocyte homolog |
| chr2_-_88553086 | 10.86 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr20_+_6869767 | 10.44 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr16_-_10802512 | 8.54 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr8_+_33239139 | 8.49 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr14_+_18983853 | 8.41 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr17_-_79085076 | 8.01 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr5_+_6373583 | 7.82 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr17_-_78499881 | 7.47 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chr14_-_103321270 | 7.43 |
ENSRNOT00000006157
|
Meis1
|
Meis homeobox 1 |
| chr13_+_42008842 | 7.36 |
ENSRNOT00000038811
|
Gpr39
|
G protein-coupled receptor 39 |
| chrM_+_9870 | 7.22 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr6_-_125723732 | 6.79 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr9_+_77320726 | 6.17 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr19_-_38484611 | 5.82 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr11_+_65743892 | 5.41 |
ENSRNOT00000085226
|
AC106292.2
|
|
| chr6_-_71199110 | 5.38 |
ENSRNOT00000081883
|
Prkd1
|
protein kinase D1 |
| chrX_+_83597265 | 4.57 |
ENSRNOT00000066667
|
LOC100911639
|
uncharacterized LOC100911639 |
| chr3_-_74868575 | 4.47 |
ENSRNOT00000051625
|
Olr544
|
olfactory receptor 544 |
| chr2_-_27287605 | 4.43 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr4_-_167068838 | 4.34 |
ENSRNOT00000029713
|
Tas2r125
|
taste receptor, type 2, member 125 |
| chr14_-_42560174 | 4.23 |
ENSRNOT00000003128
|
Tmem33
|
transmembrane protein 33 |
| chr13_+_98231326 | 4.00 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chrM_-_14061 | 3.95 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr2_+_55747318 | 3.76 |
ENSRNOT00000050655
|
Dab2
|
DAB2, clathrin adaptor protein |
| chrX_+_83678339 | 3.59 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chr5_+_116420690 | 3.41 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr18_-_28017925 | 3.24 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr8_-_53277859 | 3.11 |
ENSRNOT00000009574
|
Htr3b
|
5-hydroxytryptamine receptor 3B |
| chr13_-_98023829 | 2.92 |
ENSRNOT00000075426
|
Kif28p
|
kinesin family member 28, pseudogene |
| chr16_+_8211420 | 2.89 |
ENSRNOT00000079609
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr9_-_63291350 | 2.41 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr19_-_56983807 | 2.16 |
ENSRNOT00000043529
|
Cox6c-ps1
|
cytochrome c oxidase subunit VIc, pseudogene |
| chr3_-_77883067 | 2.15 |
ENSRNOT00000087979
|
Olr677
|
olfactory receptor 677 |
| chrM_+_2740 | 2.12 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chrX_+_152999016 | 1.90 |
ENSRNOT00000088234
|
Zfp185
|
zinc finger protein 185 |
| chr3_-_77389521 | 1.87 |
ENSRNOT00000043928
|
Olr657
|
olfactory receptor 657 |
| chr3_-_77792014 | 1.78 |
ENSRNOT00000091085
|
Olr671
|
olfactory receptor 671 |
| chr1_-_813517 | 1.71 |
ENSRNOT00000041332
|
Vom2r5
|
vomeronasal 2 receptor, 5 |
| chr1_-_173456488 | 1.63 |
ENSRNOT00000044753
|
Olr282
|
olfactory receptor 282 |
| chr10_+_87832743 | 1.61 |
ENSRNOT00000055286
|
Krtap31-1
|
keratin associated protein 31-1 |
| chr11_+_43194348 | 1.56 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr2_+_115074344 | 1.55 |
ENSRNOT00000073657
|
LOC100910500
|
olfactory receptor 144-like |
| chr1_-_134871167 | 1.48 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr1_-_205030567 | 1.46 |
ENSRNOT00000023404
|
Ctbp2
|
C-terminal binding protein 2 |
| chrM_+_14136 | 1.38 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr4_-_165774126 | 1.26 |
ENSRNOT00000007453
|
Tas2r107
|
taste receptor, type 2, member 107 |
| chr8_-_126390801 | 1.05 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr3_+_104816987 | 1.04 |
ENSRNOT00000042103
ENSRNOT00000044625 |
Fmn1
|
formin 1 |
| chr3_+_102859815 | 0.87 |
ENSRNOT00000037906
|
Olr771
|
olfactory receptor 771 |
| chr6_-_146861787 | 0.78 |
ENSRNOT00000074197
|
Abcb5
|
ATP binding cassette subfamily B member 5 |
| chr7_-_144837395 | 0.70 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr11_-_45616429 | 0.67 |
ENSRNOT00000046587
|
Olr1533
|
olfactory receptor 1533 |
| chr2_-_242828629 | 0.48 |
ENSRNOT00000043369
|
LOC499715
|
LRRGT00095 |
| chr1_-_149787308 | 0.44 |
ENSRNOT00000088514
|
LOC100909966
|
olfactory receptor 6F1-like |
| chr11_-_768644 | 0.40 |
ENSRNOT00000090197
|
Epha3
|
Eph receptor A3 |
| chr6_-_86223052 | 0.22 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr20_+_37587687 | 0.21 |
ENSRNOT00000001050
|
Msl3l2
|
male-specific lethal 3-like 2 (Drosophila) |
| chr11_-_13999862 | 0.08 |
ENSRNOT00000045835
|
Lipi
|
lipase I |
| chrX_-_35777243 | 0.03 |
ENSRNOT00000040912
|
Rs1
|
retinoschisin 1 |
| chr1_-_252167752 | 0.03 |
ENSRNOT00000074115
|
LOC100360690
|
tear acid-lipase-like protein-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfhx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.5 | 70.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 14.8 | 44.3 | GO:0010034 | response to acetate(GO:0010034) |
| 10.4 | 31.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 9.7 | 57.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 9.5 | 28.5 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 9.3 | 55.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 8.7 | 43.6 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 7.2 | 71.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 6.4 | 25.6 | GO:0051958 | methotrexate transport(GO:0051958) |
| 5.5 | 21.9 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 4.7 | 37.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 4.3 | 47.5 | GO:0015747 | urate transport(GO:0015747) |
| 3.8 | 23.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 3.3 | 13.1 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 3.0 | 26.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 2.9 | 23.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 2.8 | 8.5 | GO:0048377 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) |
| 2.5 | 7.4 | GO:0035483 | gastric emptying(GO:0035483) |
| 2.2 | 17.7 | GO:0071000 | response to magnetism(GO:0071000) |
| 2.1 | 6.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.6 | 15.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 1.3 | 3.8 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.2 | 47.3 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 1.1 | 4.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.9 | 36.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.9 | 25.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.8 | 6.8 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.6 | 19.5 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.4 | 3.2 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.3 | 7.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 4.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 55.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.3 | 1.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 0.8 | GO:0048749 | compound eye development(GO:0048749) |
| 0.3 | 3.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 1.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 5.8 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 3.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 3.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 1.4 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.2 | 50.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 7.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 2.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.1 | 1.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 11.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 7.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 4.0 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 3.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 2.9 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 2.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.4 | 31.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 8.3 | 57.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.8 | 55.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 1.6 | 13.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.4 | 17.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.4 | 6.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.2 | 6.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 1.1 | 31.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.1 | 11.6 | GO:0002177 | manchette(GO:0002177) |
| 0.9 | 3.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.9 | 25.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.9 | 23.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.8 | 4.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.6 | 76.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.5 | 3.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 36.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 5.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 82.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 14.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 157.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 8.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 10.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 8.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 66.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 6.9 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.5 | 70.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 19.3 | 57.9 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 14.4 | 71.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 12.5 | 37.6 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 9.5 | 28.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 9.3 | 55.7 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 6.5 | 19.5 | GO:0047708 | biotinidase activity(GO:0047708) |
| 6.4 | 25.6 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 5.5 | 21.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 5.3 | 31.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 4.8 | 43.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 4.8 | 48.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 3.3 | 23.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.7 | 54.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 2.6 | 13.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 2.2 | 17.7 | GO:0071253 | connexin binding(GO:0071253) |
| 1.4 | 8.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.9 | 12.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.9 | 26.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.6 | 47.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.5 | 25.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.5 | 12.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 50.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 11.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.4 | 35.8 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.4 | 3.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 5.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 3.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 4.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 25.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 25.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 3.1 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 1.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 2.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 1.5 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 6.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 15.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 50.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 1.3 | 71.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.8 | 31.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.6 | 43.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.5 | 32.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.5 | 23.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.5 | 17.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 5.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 5.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 51.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 8.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 8.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 17.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 6.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 55.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 3.2 | 55.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 3.1 | 106.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.6 | 31.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.5 | 71.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 1.4 | 26.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.3 | 17.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.2 | 32.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.1 | 12.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 3.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 8.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 24.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 37.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 5.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 7.3 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |