Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
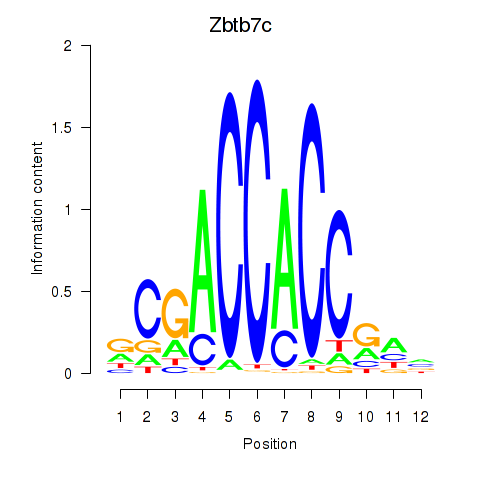
Results for Zbtb7c
Z-value: 0.63
Transcription factors associated with Zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7c
|
ENSRNOG00000047924 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7c | rn6_v1_chr18_+_72005581_72005581 | -0.08 | 1.8e-01 | Click! |
Activity profile of Zbtb7c motif
Sorted Z-values of Zbtb7c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88122233 | 74.84 |
ENSRNOT00000083895
ENSRNOT00000005285 |
Krt14
|
keratin 14 |
| chr10_-_88050622 | 55.75 |
ENSRNOT00000019037
|
Krt15
|
keratin 15 |
| chr5_+_152681101 | 23.00 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr15_+_104095179 | 20.15 |
ENSRNOT00000093487
|
Cldn10
|
claudin 10 |
| chr1_+_224824799 | 9.31 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr3_+_172856733 | 7.50 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr3_+_154043873 | 6.51 |
ENSRNOT00000072502
ENSRNOT00000034166 |
Nnat
|
neuronatin |
| chr4_-_184096806 | 6.05 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr1_+_104576589 | 5.23 |
ENSRNOT00000046529
|
Nav2
|
neuron navigator 2 |
| chr10_+_55013703 | 4.99 |
ENSRNOT00000032785
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chrX_+_15049462 | 4.41 |
ENSRNOT00000007015
|
Ebp
|
emopamil binding protein (sterol isomerase) |
| chr16_+_9563218 | 3.84 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr10_+_92289107 | 3.82 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr12_-_37425596 | 3.16 |
ENSRNOT00000084553
|
Tctn2
|
tectonic family member 2 |
| chr1_-_259106948 | 3.05 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr5_-_127983515 | 3.03 |
ENSRNOT00000043054
|
Fam159a
|
family with sequence similarity 159, member A |
| chrX_-_124766044 | 3.01 |
ENSRNOT00000000177
|
Lamp2
|
lysosomal-associated membrane protein 2 |
| chr15_-_34693034 | 2.98 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr3_+_146582752 | 2.96 |
ENSRNOT00000010157
|
Pygb
|
glycogen phosphorylase B |
| chr10_+_57185347 | 2.71 |
ENSRNOT00000040488
ENSRNOT00000086374 |
Mink1
|
misshapen-like kinase 1 |
| chr17_-_44420596 | 2.67 |
ENSRNOT00000077512
|
RGD1561897
|
similar to Serine/threonine-protein kinase Kist (Kinase interacting with stathmin) |
| chr13_-_88536728 | 2.65 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chrX_-_158739838 | 2.57 |
ENSRNOT00000003091
|
Phf6
|
PHD finger protein 6 |
| chr19_+_25969255 | 2.46 |
ENSRNOT00000004370
|
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_+_169590308 | 2.45 |
ENSRNOT00000023147
|
Olr155
|
olfactory receptor 155 |
| chr7_+_53878610 | 2.39 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr10_+_59360765 | 2.36 |
ENSRNOT00000036278
|
Zzef1
|
zinc finger ZZ-type and EF-hand domain containing 1 |
| chr5_+_22392732 | 2.24 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr20_-_6961162 | 2.18 |
ENSRNOT00000000635
|
Mtch1
|
mitochondrial carrier 1 |
| chr5_-_139921013 | 2.07 |
ENSRNOT00000079957
|
Smap2
|
small ArfGAP2 |
| chr8_+_73682887 | 1.90 |
ENSRNOT00000057522
|
Vps13c
|
vacuolar protein sorting 13C |
| chr20_-_6960481 | 1.73 |
ENSRNOT00000093172
|
Mtch1
|
mitochondrial carrier 1 |
| chrX_-_128268285 | 1.68 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr5_+_26493212 | 1.60 |
ENSRNOT00000061328
|
Triqk
|
triple QxxK/R motif containing |
| chr5_+_142845114 | 1.57 |
ENSRNOT00000039870
|
Yrdc
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr10_+_20320878 | 1.57 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr10_+_65448950 | 1.26 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr4_-_132740938 | 1.21 |
ENSRNOT00000007185
|
Rybp
|
RING1 and YY1 binding protein |
| chr13_-_72367980 | 1.15 |
ENSRNOT00000003928
|
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr12_+_41543696 | 1.14 |
ENSRNOT00000079639
|
Tpcn1
|
two pore segment channel 1 |
| chr10_+_104118945 | 1.09 |
ENSRNOT00000005035
|
Nup85
|
nucleoporin 85 |
| chr1_-_214844858 | 1.04 |
ENSRNOT00000046344
|
Tollip
|
toll interacting protein |
| chr2_-_24923128 | 0.87 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr8_+_72029489 | 0.76 |
ENSRNOT00000089336
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr14_+_38192870 | 0.71 |
ENSRNOT00000077080
|
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr3_-_72445775 | 0.69 |
ENSRNOT00000033427
ENSRNOT00000077573 |
P2rx3
|
purinergic receptor P2X 3 |
| chr10_-_59360661 | 0.65 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr4_+_176994129 | 0.44 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr12_+_21981755 | 0.39 |
ENSRNOT00000001871
|
LOC100910540
|
7SK snRNA methylphosphate capping enzyme-like |
| chr3_-_113231790 | 0.30 |
ENSRNOT00000019025
|
Tp53bp1
|
tumor protein p53 binding protein 1 |
| chr2_-_198852161 | 0.25 |
ENSRNOT00000028815
|
Polr3c
|
RNA polymerase III subunit C |
| chr9_-_43022998 | 0.18 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr5_-_36683356 | 0.07 |
ENSRNOT00000009043
|
Pou3f2
|
POU class 3 homeobox 2 |
| chr4_+_133285552 | 0.04 |
ENSRNOT00000029115
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
| chr3_-_95418679 | 0.02 |
ENSRNOT00000018074
|
Rcn1
|
reticulocalbin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 74.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 5.7 | 23.0 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.9 | 7.5 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 1.6 | 9.3 | GO:0031427 | response to methotrexate(GO:0031427) |
| 1.3 | 5.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.1 | 53.9 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 1.0 | 3.0 | GO:1905146 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) lysosomal protein catabolic process(GO:1905146) |
| 0.6 | 2.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.5 | 1.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.5 | 2.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 | 3.8 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) regulation of cellular response to heat(GO:1900034) |
| 0.5 | 3.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 1.6 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.3 | 1.9 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.2 | 0.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 2.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 1.7 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 1.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 1.3 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 3.0 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 2.7 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 5.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 4.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 1.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 6.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 3.9 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 2.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.1 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 3.0 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 128.8 | GO:0045095 | keratin filament(GO:0045095) |
| 1.7 | 5.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 1.0 | 3.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) lysosomal matrix(GO:1990836) |
| 1.0 | 3.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.8 | 2.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 2.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 5.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 1.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 20.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 9.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 19.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 4.3 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.0 | 74.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.5 | 7.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 2.0 | 6.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 1.5 | 4.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 1.2 | 9.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.0 | 3.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 1.0 | 3.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.8 | 5.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.6 | 54.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.6 | 1.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.4 | 2.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 2.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 1.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 1.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.0 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 2.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 23.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 1.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 3.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 5.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.7 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 26.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.1 | 72.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 7.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 4.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.0 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.6 | 20.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 5.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 3.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 7.5 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |