Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zbtb7b
Z-value: 0.76
Transcription factors associated with Zbtb7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7b
|
ENSRNOG00000020640 | zinc finger and BTB domain containing 7B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7b | rn6_v1_chr2_-_188718704_188718704 | 0.15 | 9.0e-03 | Click! |
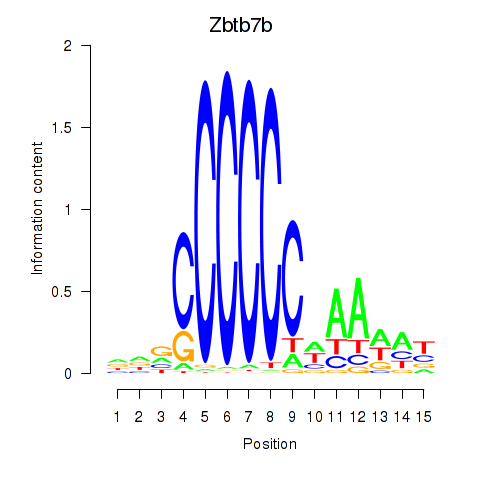
Activity profile of Zbtb7b motif
Sorted Z-values of Zbtb7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_255376833 | 25.88 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr18_-_15540177 | 20.91 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr15_-_28313841 | 19.57 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr1_-_215834704 | 19.57 |
ENSRNOT00000073850
|
Igf2
|
insulin-like growth factor 2 |
| chr10_+_39655455 | 18.64 |
ENSRNOT00000058817
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr4_-_4473307 | 17.47 |
ENSRNOT00000045773
|
Dpp6
|
dipeptidyl peptidase like 6 |
| chr15_+_33600102 | 17.12 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr17_-_69827112 | 15.58 |
ENSRNOT00000023835
|
Akr1c14
|
aldo-keto reductase family 1, member C14 |
| chr3_+_92640752 | 15.20 |
ENSRNOT00000007604
|
Slc1a2
|
solute carrier family 1 member 2 |
| chr10_+_86657285 | 14.98 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chrX_-_142248369 | 14.59 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr14_-_92117923 | 14.25 |
ENSRNOT00000044726
|
Grb10
|
growth factor receptor bound protein 10 |
| chr2_-_179704629 | 13.96 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr8_-_98738446 | 13.59 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr17_+_12762752 | 13.28 |
ENSRNOT00000044842
|
Diras2
|
DIRAS family GTPase 2 |
| chrX_+_110016995 | 13.14 |
ENSRNOT00000093542
|
Nrk
|
Nik related kinase |
| chr1_-_80599572 | 13.02 |
ENSRNOT00000024832
|
Apoc4
|
apolipoprotein C4 |
| chr4_-_125808281 | 12.85 |
ENSRNOT00000037848
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr15_+_33600337 | 12.35 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr18_-_64177729 | 12.18 |
ENSRNOT00000022347
|
Mc2r
|
melanocortin 2 receptor |
| chr7_-_11018160 | 11.98 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr4_-_179307095 | 11.97 |
ENSRNOT00000021193
|
Bcat1
|
branched chain amino acid transaminase 1 |
| chr8_+_74868681 | 11.72 |
ENSRNOT00000039376
|
AABR07070555.1
|
|
| chr14_+_12218553 | 11.72 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr20_-_4823475 | 11.50 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chrX_-_152642531 | 11.30 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr5_+_139783951 | 10.96 |
ENSRNOT00000081333
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr7_-_138483612 | 10.96 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr8_+_53411316 | 10.87 |
ENSRNOT00000011107
ENSRNOT00000086957 |
Tmprss5
|
transmembrane protease, serine 5 |
| chr3_-_48535909 | 10.51 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr17_-_10766253 | 10.19 |
ENSRNOT00000000117
|
Cplx2
|
complexin 2 |
| chr3_+_63379031 | 9.99 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr1_-_222246765 | 9.86 |
ENSRNOT00000087285
ENSRNOT00000028731 |
Dnajc4
|
DnaJ heat shock protein family (Hsp40) member C4 |
| chr2_+_121165137 | 9.75 |
ENSRNOT00000016236
|
Sox2
|
SRY box 2 |
| chr5_-_2982603 | 9.64 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr3_+_56056925 | 9.58 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr8_-_39460844 | 9.24 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr12_-_42492328 | 9.12 |
ENSRNOT00000011552
|
Tbx3
|
T-box 3 |
| chr8_-_23063041 | 9.09 |
ENSRNOT00000018416
|
Elavl3
|
ELAV like RNA binding protein 3 |
| chr3_-_176459533 | 8.84 |
ENSRNOT00000079283
|
Nkain4
|
Sodium/potassium transporting ATPase interacting 4 |
| chr8_-_120446455 | 8.80 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr20_-_5020150 | 8.76 |
ENSRNOT00000001146
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr7_+_78131232 | 8.74 |
ENSRNOT00000039984
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr2_+_144861455 | 8.65 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr10_-_27366665 | 8.57 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr3_+_172385672 | 8.49 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr10_-_36323055 | 8.30 |
ENSRNOT00000081026
|
Zfp354c
|
zinc finger protein 354C |
| chr8_+_117836701 | 8.17 |
ENSRNOT00000043345
|
Plxnb1
|
plexin B1 |
| chr19_-_37427989 | 8.16 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr10_-_36322356 | 8.06 |
ENSRNOT00000000248
|
Zfp354c
|
zinc finger protein 354C |
| chr1_-_89488223 | 7.88 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chrX_-_1704033 | 7.73 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chrX_-_13279082 | 7.67 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr13_+_51034256 | 7.45 |
ENSRNOT00000004528
ENSRNOT00000046854 ENSRNOT00000087320 |
Mybph
|
myosin binding protein H |
| chr1_-_198232344 | 7.35 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr12_-_42492526 | 7.27 |
ENSRNOT00000084018
|
Tbx3
|
T-box 3 |
| chr10_+_69737328 | 7.04 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr3_+_95711555 | 6.79 |
ENSRNOT00000006302
|
Pax6
|
paired box 6 |
| chr11_+_86797557 | 6.54 |
ENSRNOT00000083049
ENSRNOT00000046594 |
Tango2
|
transport and golgi organization 2 homolog |
| chr16_-_19872774 | 6.50 |
ENSRNOT00000000060
|
Abhd8
|
abhydrolase domain containing 8 |
| chr19_+_60104804 | 6.49 |
ENSRNOT00000078559
|
Pard3
|
par-3 family cell polarity regulator |
| chr19_+_25095089 | 6.26 |
ENSRNOT00000041717
|
Prkaca
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr6_+_126018841 | 6.11 |
ENSRNOT00000089840
ENSRNOT00000088089 |
Slc24a4
|
solute carrier family 24 member 4 |
| chr1_+_213870502 | 6.04 |
ENSRNOT00000086483
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr6_-_104409005 | 5.96 |
ENSRNOT00000040965
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr10_+_56627411 | 5.88 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr8_-_77398156 | 5.81 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
| chr10_-_86004096 | 5.74 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr1_-_72335855 | 5.66 |
ENSRNOT00000021613
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr1_-_164307084 | 5.63 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr3_-_44522930 | 5.60 |
ENSRNOT00000006963
|
Acvr1
|
activin A receptor type 1 |
| chr20_+_20105047 | 5.34 |
ENSRNOT00000082181
|
Ank3
|
ankyrin 3 |
| chr1_-_101085884 | 5.18 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr11_-_87081950 | 5.17 |
ENSRNOT00000002574
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr19_+_387374 | 5.16 |
ENSRNOT00000075433
|
LOC102547811
|
uncharacterized LOC102547811 |
| chr20_-_29579578 | 5.13 |
ENSRNOT00000000700
|
AC096404.1
|
|
| chr3_-_111469518 | 5.13 |
ENSRNOT00000006874
|
Ndufaf1
|
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
| chr8_-_36410612 | 4.91 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr1_-_80615704 | 4.81 |
ENSRNOT00000041891
|
Apoe
|
apolipoprotein E |
| chr12_+_18531990 | 4.69 |
ENSRNOT00000036606
|
Asmtl
|
acetylserotonin O-methyltransferase-like |
| chr2_+_60180215 | 4.62 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr2_+_140541619 | 4.60 |
ENSRNOT00000017396
|
Rab33b
|
RAB33B, member RAS oncogene family |
| chr18_-_5314511 | 4.58 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr2_+_193275507 | 4.56 |
ENSRNOT00000091537
|
LOC100361951
|
hypothetical protein LOC100361951 |
| chr12_+_18516946 | 4.43 |
ENSRNOT00000029485
|
Dhrsx
|
dehydrogenase/reductase X-linked |
| chr10_+_66099531 | 4.43 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr10_-_103873246 | 4.43 |
ENSRNOT00000039387
|
Ush1g
|
Usher syndrome 1G |
| chr4_+_121760773 | 4.38 |
ENSRNOT00000087069
|
Vom1r92
|
vomeronasal 1 receptor 92 |
| chr10_+_88914276 | 4.38 |
ENSRNOT00000087076
ENSRNOT00000055238 |
Atp6v0a1
|
ATPase H+ transporting V0 subunit a1 |
| chr2_+_85377318 | 4.30 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr1_-_80615525 | 4.28 |
ENSRNOT00000091574
|
Apoe
|
apolipoprotein E |
| chr15_+_33755478 | 4.23 |
ENSRNOT00000034073
|
Thtpa
|
thiamine triphosphatase |
| chr9_-_65916949 | 4.10 |
ENSRNOT00000077483
|
Tmem237
|
transmembrane protein 237 |
| chr20_-_422464 | 4.09 |
ENSRNOT00000051646
|
Olr1673
|
olfactory receptor 1673 |
| chrX_+_77065397 | 4.06 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr1_-_189911571 | 4.04 |
ENSRNOT00000080996
ENSRNOT00000088536 |
Zp2
|
zona pellucida glycoprotein 2 |
| chr1_-_173393390 | 4.00 |
ENSRNOT00000050657
|
Olr285
|
olfactory receptor 285 |
| chr3_-_2719513 | 3.98 |
ENSRNOT00000020997
|
Lcn12
|
lipocalin 12 |
| chr7_+_6644643 | 3.93 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr13_-_89668473 | 3.85 |
ENSRNOT00000004938
|
Ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr1_+_255185629 | 3.85 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr15_+_47373120 | 3.82 |
ENSRNOT00000070815
|
Rp1l1
|
RP1 like 1 |
| chr9_-_65917132 | 3.73 |
ENSRNOT00000031866
|
Tmem237
|
transmembrane protein 237 |
| chr11_-_62396909 | 3.70 |
ENSRNOT00000093596
ENSRNOT00000093179 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_-_106695570 | 3.66 |
ENSRNOT00000083517
|
Hhla1
|
HERV-H LTR-associating 1 |
| chr1_+_64101018 | 3.64 |
ENSRNOT00000078640
|
Mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr20_+_25990656 | 3.63 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr19_-_25366806 | 3.61 |
ENSRNOT00000003762
|
Mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr8_+_129205931 | 3.60 |
ENSRNOT00000082377
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr3_-_2719135 | 3.59 |
ENSRNOT00000080257
|
Lcn12
|
lipocalin 12 |
| chr20_+_5455974 | 3.58 |
ENSRNOT00000000553
ENSRNOT00000092676 |
Pfdn6
|
prefoldin subunit 6 |
| chr3_-_23020441 | 3.56 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr13_+_48689962 | 3.54 |
ENSRNOT00000079707
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr14_-_18076907 | 3.50 |
ENSRNOT00000003472
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr18_-_63357194 | 3.47 |
ENSRNOT00000089408
ENSRNOT00000066103 |
Spire1
|
spire-type actin nucleation factor 1 |
| chr1_+_84612448 | 3.44 |
ENSRNOT00000032991
|
AABR07071891.1
|
|
| chrX_-_110230610 | 3.33 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chr3_-_160465143 | 3.33 |
ENSRNOT00000040156
|
Tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr1_+_83714347 | 3.30 |
ENSRNOT00000085245
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr10_-_94406949 | 3.28 |
ENSRNOT00000012533
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr9_+_3896337 | 3.25 |
ENSRNOT00000079166
|
LOC103693189
|
protein tyrosine phosphatase type IVA 1 |
| chr2_-_184951950 | 3.24 |
ENSRNOT00000093486
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr15_-_5894854 | 3.23 |
ENSRNOT00000024427
|
Cd99l2
|
CD99 molecule-like 2 |
| chr1_-_261669584 | 3.22 |
ENSRNOT00000020568
ENSRNOT00000076555 |
Crtac1
|
cartilage acidic protein 1 |
| chr15_+_5265307 | 3.16 |
ENSRNOT00000031807
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr12_+_38375416 | 3.14 |
ENSRNOT00000078530
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr7_-_2623781 | 3.04 |
ENSRNOT00000004173
|
Spryd4
|
SPRY domain containing 4 |
| chr16_-_20641908 | 3.02 |
ENSRNOT00000026846
|
Ell
|
elongation factor for RNA polymerase II |
| chr10_-_14703668 | 2.94 |
ENSRNOT00000024767
|
Tpsab1
|
tryptase alpha/beta 1 |
| chr3_-_107760550 | 2.94 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr10_+_90550147 | 2.89 |
ENSRNOT00000032944
|
Fzd2
|
frizzled class receptor 2 |
| chr2_-_105224295 | 2.89 |
ENSRNOT00000014178
|
Agtr1b
|
angiotensin II receptor, type 1b |
| chr7_-_16068823 | 2.87 |
ENSRNOT00000073630
|
Olr922
|
olfactory receptor 922 |
| chr3_+_100788806 | 2.82 |
ENSRNOT00000083289
ENSRNOT00000090445 |
Bdnf
|
brain-derived neurotrophic factor |
| chr8_+_122003916 | 2.78 |
ENSRNOT00000091980
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr4_+_85427555 | 2.76 |
ENSRNOT00000015469
|
Fam188b
|
family with sequence similarity 188, member B |
| chr1_+_276309927 | 2.69 |
ENSRNOT00000067460
ENSRNOT00000066236 |
Vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr7_+_121311024 | 2.69 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr1_-_88780425 | 2.67 |
ENSRNOT00000074494
|
Sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr20_+_5456235 | 2.65 |
ENSRNOT00000092555
|
Pfdn6
|
prefoldin subunit 6 |
| chr7_-_116063078 | 2.55 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
| chr18_-_80865584 | 2.52 |
ENSRNOT00000021750
|
Tshz1
|
teashirt zinc finger homeobox 1 |
| chr12_+_2046472 | 2.48 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr19_-_36157924 | 2.47 |
ENSRNOT00000072022
|
AABR07043701.1
|
|
| chr6_+_129538982 | 2.44 |
ENSRNOT00000083626
ENSRNOT00000082522 |
Ak7
|
adenylate kinase 7 |
| chr1_+_215460226 | 2.43 |
ENSRNOT00000027270
|
LOC685544
|
hypothetical protein LOC685544 |
| chr19_+_85606 | 2.42 |
ENSRNOT00000015724
|
Ces2e
|
carboxylesterase 2E |
| chr10_+_70520206 | 2.39 |
ENSRNOT00000088198
ENSRNOT00000090446 ENSRNOT00000085799 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr1_-_225329770 | 2.37 |
ENSRNOT00000027459
|
Asrgl1
|
asparaginase like 1 |
| chr10_-_87578854 | 2.30 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chr5_+_58144705 | 2.23 |
ENSRNOT00000019886
|
Galt
|
galactose-1-phosphate uridylyltransferase |
| chr20_-_35342 | 2.20 |
ENSRNOT00000048277
|
LOC100910263
|
olfactory receptor 2B11-like |
| chr16_-_81583916 | 2.18 |
ENSRNOT00000026281
|
LOC103693999
|
transmembrane and coiled-coil domain-containing protein 3 |
| chr12_+_4248808 | 2.18 |
ENSRNOT00000042410
|
AABR07035089.1
|
|
| chr2_-_198719202 | 2.12 |
ENSRNOT00000028801
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr11_+_30363280 | 2.11 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr3_-_77525518 | 2.11 |
ENSRNOT00000049513
|
Olr663
|
olfactory receptor 663 |
| chr12_-_39641285 | 2.08 |
ENSRNOT00000001729
|
Anapc7
|
anaphase promoting complex subunit 7 |
| chr19_+_37330930 | 2.02 |
ENSRNOT00000022439
|
Plekhg4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr9_-_104478959 | 1.98 |
ENSRNOT00000034533
|
AABR07068417.1
|
|
| chr4_+_121899292 | 1.94 |
ENSRNOT00000077949
|
Vom1r97
|
vomeronasal 1 receptor 97 |
| chr4_-_55740627 | 1.92 |
ENSRNOT00000010658
|
Pax4
|
paired box 4 |
| chr12_+_22229079 | 1.90 |
ENSRNOT00000001911
|
Gnb2
|
G protein subunit beta 2 |
| chr5_+_156618962 | 1.87 |
ENSRNOT00000020069
|
Sh2d5
|
SH2 domain containing 5 |
| chr14_-_65153013 | 1.85 |
ENSRNOT00000071542
|
AABR07015596.1
|
|
| chr3_-_6017597 | 1.82 |
ENSRNOT00000065867
|
Brd3
|
bromodomain containing 3 |
| chr8_+_19972793 | 1.80 |
ENSRNOT00000068066
|
Olr1159
|
olfactory receptor 1159 |
| chr1_+_86948918 | 1.78 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr10_+_105235289 | 1.73 |
ENSRNOT00000075548
|
Galr2
|
galanin receptor 2 |
| chr3_+_8547349 | 1.68 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr2_+_186630278 | 1.67 |
ENSRNOT00000021604
|
LOC365839
|
similar to elongation protein 4 homolog |
| chr16_-_2226121 | 1.62 |
ENSRNOT00000091513
|
Slmap
|
sarcolemma associated protein |
| chr1_-_185569190 | 1.62 |
ENSRNOT00000090773
|
RGD1311703
|
similar to sid2057p |
| chr6_-_108167185 | 1.61 |
ENSRNOT00000015545
|
Aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_-_72339395 | 1.60 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr4_+_157057601 | 1.58 |
ENSRNOT00000040592
|
Zfp42l
|
Zinc finger protein 42-like |
| chr20_+_4823662 | 1.56 |
ENSRNOT00000090028
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr16_-_75028977 | 1.55 |
ENSRNOT00000058071
|
Defb13
|
defensin beta 13 |
| chr1_+_84009268 | 1.54 |
ENSRNOT00000057230
ENSRNOT00000081121 |
RGD1560854
|
similar to FLJ41131 protein |
| chr1_-_65659945 | 1.54 |
ENSRNOT00000058574
|
Rnf225
|
ring finger protein 225 |
| chr1_-_221431713 | 1.53 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr1_+_79898551 | 1.49 |
ENSRNOT00000019069
|
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr17_-_32953641 | 1.46 |
ENSRNOT00000023332
|
Wrnip1
|
Werner helicase interacting protein 1 |
| chr8_+_64531594 | 1.43 |
ENSRNOT00000036798
|
Gramd2
|
GRAM domain containing 2 |
| chr10_+_14105750 | 1.42 |
ENSRNOT00000090552
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chr15_-_60289763 | 1.32 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr4_+_140247313 | 1.31 |
ENSRNOT00000040255
ENSRNOT00000064025 ENSRNOT00000041130 ENSRNOT00000043646 |
Itpr1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr1_+_100593892 | 1.26 |
ENSRNOT00000027062
|
Kcnc3
|
potassium voltage-gated channel subfamily C member 3 |
| chr11_-_23063414 | 1.26 |
ENSRNOT00000043396
|
LOC103690036
|
olfactory receptor 6C6-like |
| chr1_+_47972399 | 1.19 |
ENSRNOT00000033408
ENSRNOT00000077707 |
Acat2
|
acetyl-CoA acetyltransferase 2 |
| chr3_+_113131327 | 1.16 |
ENSRNOT00000018460
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr20_-_10257044 | 1.16 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr5_-_137321121 | 1.15 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chrX_+_21499934 | 1.13 |
ENSRNOT00000090890
ENSRNOT00000081037 |
Huwe1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr10_-_14072230 | 1.10 |
ENSRNOT00000018405
|
Tbl3
|
transducin (beta)-like 3 |
| chr5_-_137901590 | 1.10 |
ENSRNOT00000044946
|
Olr868
|
olfactory receptor 868 |
| chrX_-_1369384 | 1.09 |
ENSRNOT00000013745
|
Timp1
|
TIMP metallopeptidase inhibitor 1 |
| chr5_-_136762986 | 1.04 |
ENSRNOT00000026852
|
Artn
|
artemin |
| chr4_-_157274755 | 1.04 |
ENSRNOT00000088895
|
LOC100911672
|
atrophin-1-like |
| chr14_-_79987915 | 0.99 |
ENSRNOT00000089006
|
Afap1
|
actin filament associated protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 5.5 | 16.4 | GO:0060932 | sinoatrial node cell development(GO:0060931) His-Purkinje system cell differentiation(GO:0060932) |
| 5.0 | 15.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 4.9 | 19.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 4.0 | 12.0 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 3.9 | 11.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 3.5 | 10.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 3.0 | 15.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 3.0 | 9.1 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 2.8 | 8.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 2.8 | 19.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 2.7 | 8.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 2.4 | 9.8 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 2.4 | 25.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 2.3 | 6.8 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.2 | 24.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.2 | 13.1 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 2.1 | 14.6 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 2.0 | 7.9 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 1.9 | 5.8 | GO:0010034 | response to acetate(GO:0010034) |
| 1.9 | 5.6 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 1.9 | 13.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.6 | 11.3 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 1.5 | 4.6 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 1.4 | 4.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.3 | 5.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.3 | 12.0 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.2 | 5.9 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 1.2 | 3.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 1.1 | 6.5 | GO:0003383 | apical constriction(GO:0003383) |
| 1.0 | 6.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 1.0 | 4.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 1.0 | 29.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 1.0 | 2.9 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 1.0 | 2.9 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.9 | 3.5 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.8 | 10.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.8 | 2.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.8 | 3.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.8 | 3.9 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.7 | 7.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 2.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.7 | 5.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.7 | 8.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.7 | 8.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.6 | 3.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.6 | 7.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.6 | 3.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.6 | 1.8 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.6 | 14.0 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.6 | 2.2 | GO:0061622 | UDP-glucose metabolic process(GO:0006011) glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.6 | 4.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.5 | 4.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.5 | 1.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.5 | 2.7 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.5 | 4.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 3.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.5 | 2.0 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.5 | 1.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 3.3 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.5 | 17.5 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.4 | 2.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 14.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 3.6 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 | 6.3 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.4 | 1.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) ketone body catabolic process(GO:0046952) |
| 0.4 | 2.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.4 | 4.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.4 | 3.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.4 | 11.0 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 1.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.3 | 1.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.3 | 6.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 1.6 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.3 | 9.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.3 | 1.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.3 | 2.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 3.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.3 | 1.1 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.3 | 2.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 4.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 2.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.5 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 1.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 3.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 8.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 0.8 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.2 | 1.6 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 1.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 12.2 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.2 | 11.9 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.2 | 15.6 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 14.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 11.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 17.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.8 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 6.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.6 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 | 4.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 8.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 3.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.7 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.9 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) |
| 0.1 | 6.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.5 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 4.9 | GO:0031016 | pancreas development(GO:0031016) |
| 0.1 | 12.3 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 4.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 1.1 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.6 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 1.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.9 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.4 | GO:0032720 | negative regulation of tumor necrosis factor production(GO:0032720) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.4 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 3.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 4.0 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 6.9 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 6.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 21.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.2 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 2.2 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 25.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 1.8 | 9.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.7 | 10.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.7 | 19.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 1.1 | 5.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 1.1 | 6.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.9 | 15.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.9 | 13.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.9 | 19.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.8 | 6.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.8 | 3.0 | GO:0035363 | histone locus body(GO:0035363) |
| 0.7 | 6.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 7.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.5 | 5.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 16.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 7.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 2.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.4 | 4.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 5.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 19.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 1.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 1.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.3 | 3.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 8.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 9.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 19.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.3 | 1.8 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 7.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 14.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.9 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.2 | 4.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 7.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 9.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 17.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 1.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.2 | 9.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 4.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 1.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 2.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 9.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 4.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 5.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 5.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 12.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 3.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 7.5 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 10.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 11.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 8.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.3 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 3.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 7.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 27.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 3.9 | 15.6 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 3.6 | 18.0 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 3.0 | 15.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 3.0 | 9.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 3.0 | 35.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 2.9 | 8.6 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.8 | 14.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.4 | 12.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 1.6 | 18.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 1.5 | 4.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.4 | 8.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.3 | 11.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.1 | 4.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.1 | 7.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.0 | 19.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.0 | 5.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 1.0 | 5.8 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.9 | 5.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.9 | 6.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.7 | 6.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 9.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.6 | 2.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.6 | 1.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.6 | 8.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.6 | 2.9 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.6 | 7.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.5 | 14.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.5 | 7.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 4.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.5 | 22.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 11.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.4 | 2.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 2.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 9.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.4 | 3.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.4 | 1.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.4 | 9.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 3.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.4 | 12.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.4 | 1.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.4 | 2.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 17.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.3 | 1.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.3 | 1.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 9.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 10.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 4.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 11.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 3.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 18.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 2.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 10.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 10.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 6.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 2.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 3.6 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.2 | 19.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 2.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 6.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 4.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 1.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 3.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 14.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 2.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 11.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 8.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 8.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 1.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 6.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 3.6 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 3.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.6 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 4.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 14.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 2.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 3.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 1.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 21.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 22.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 20.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.4 | 20.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 5.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 14.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 15.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 12.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 10.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 13.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 10.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 8.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 8.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 15.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 2.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 5.7 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 19.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.0 | 18.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 1.0 | 11.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.9 | 14.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.9 | 19.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 16.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.8 | 13.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 13.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.5 | 20.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 11.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 26.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.4 | 10.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.4 | 6.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.3 | 6.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 6.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 18.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 7.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 4.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 7.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 8.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 3.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 10.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 4.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 4.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 3.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 6.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 13.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 4.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |