Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
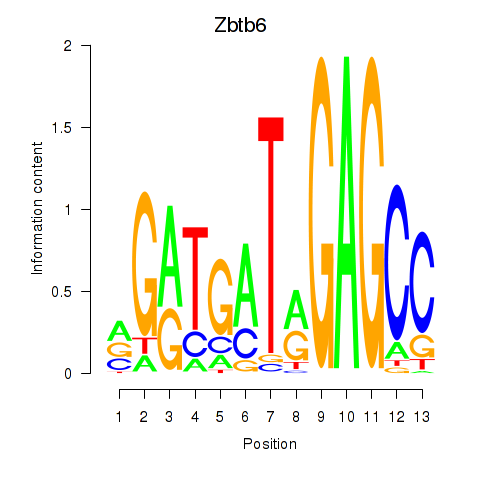
Results for Zbtb6
Z-value: 0.98
Transcription factors associated with Zbtb6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb6
|
ENSRNOG00000009340 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb6 | rn6_v1_chr3_-_21684132_21684132 | 0.27 | 6.2e-07 | Click! |
Activity profile of Zbtb6 motif
Sorted Z-values of Zbtb6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_105279462 | 53.16 |
ENSRNOT00000010679
|
Scg5
|
secretogranin V |
| chr10_-_103816287 | 49.02 |
ENSRNOT00000004477
|
Grin2c
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr1_-_31122093 | 46.33 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr10_-_90999506 | 45.22 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr17_+_88215834 | 42.96 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr10_-_27366665 | 42.37 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr2_+_174013288 | 29.51 |
ENSRNOT00000013904
|
Serpini1
|
serpin family I member 1 |
| chr12_-_23925741 | 28.85 |
ENSRNOT00000001957
|
Srrm3
|
serine/arginine repetitive matrix 3 |
| chr16_-_18937562 | 27.22 |
ENSRNOT00000080387
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr7_+_78092037 | 26.93 |
ENSRNOT00000050753
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr7_+_145117951 | 25.24 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr8_+_48382121 | 24.34 |
ENSRNOT00000008685
|
Thy1
|
Thy-1 cell surface antigen |
| chrX_+_114929029 | 24.08 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr10_-_51669297 | 23.78 |
ENSRNOT00000071595
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr14_+_25589762 | 22.56 |
ENSRNOT00000043938
ENSRNOT00000067439 ENSRNOT00000002793 |
Epha5
|
EPH receptor A5 |
| chr1_+_214927172 | 22.41 |
ENSRNOT00000027134
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr8_-_98738446 | 22.24 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr18_+_30435119 | 21.80 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr10_-_89958805 | 21.38 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr3_-_94182714 | 21.17 |
ENSRNOT00000015073
|
LOC100362814
|
hypothetical protein LOC100362814 |
| chrX_-_115764217 | 20.16 |
ENSRNOT00000009400
|
Trpc5
|
transient receptor potential cation channel, subfamily C, member 5 |
| chr3_+_172374957 | 17.38 |
ENSRNOT00000035916
|
Gnas
|
GNAS complex locus |
| chr3_-_15278645 | 16.95 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr13_+_51022681 | 16.94 |
ENSRNOT00000078599
|
Chi3l1
|
chitinase 3 like 1 |
| chr20_-_53777159 | 16.65 |
ENSRNOT00000073994
|
AABR07045621.1
|
|
| chr10_+_103308392 | 16.59 |
ENSRNOT00000004141
|
Kif19
|
kinesin family member 19 |
| chr5_-_62621737 | 16.46 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr2_+_3400977 | 16.27 |
ENSRNOT00000093593
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr11_+_87366621 | 15.93 |
ENSRNOT00000040978
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr15_-_33576331 | 15.68 |
ENSRNOT00000021851
|
Slc22a17
|
solute carrier family 22, member 17 |
| chr18_-_56331991 | 15.04 |
ENSRNOT00000085841
ENSRNOT00000091220 |
Slc6a7
|
solute carrier family 6 member 7 |
| chr7_+_23403891 | 15.02 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr13_+_71086745 | 14.80 |
ENSRNOT00000083366
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr14_-_114484127 | 14.79 |
ENSRNOT00000056706
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr17_-_48562838 | 14.27 |
ENSRNOT00000017102
ENSRNOT00000084702 |
Amph
|
amphiphysin |
| chrX_+_2435305 | 13.95 |
ENSRNOT00000005623
|
Slc9a7
|
solute carrier family 9 member A7 |
| chr18_+_52917124 | 13.64 |
ENSRNOT00000021921
|
Slc12a2
|
solute carrier family 12 member 2 |
| chr1_+_172752416 | 13.51 |
ENSRNOT00000087448
|
Olr268
|
olfactory receptor 268 |
| chr1_+_221801524 | 13.37 |
ENSRNOT00000031227
|
Nrxn2
|
neurexin 2 |
| chr1_-_220136470 | 12.29 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chrX_-_22440187 | 12.25 |
ENSRNOT00000090731
|
Gpr173
|
|
| chr3_-_175479395 | 12.16 |
ENSRNOT00000077308
|
Hrh3
|
histamine receptor H3 |
| chr8_+_24369916 | 12.11 |
ENSRNOT00000033460
|
Bmper
|
BMP-binding endothelial regulator |
| chr19_-_58735173 | 11.98 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr1_+_221792221 | 11.68 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr2_+_41911131 | 11.16 |
ENSRNOT00000016768
|
Plk2
|
polo-like kinase 2 |
| chr3_+_92403582 | 11.08 |
ENSRNOT00000064282
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr8_+_122003916 | 10.77 |
ENSRNOT00000091980
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr7_-_70480724 | 10.40 |
ENSRNOT00000090057
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr5_+_123905166 | 10.37 |
ENSRNOT00000082021
|
Dab1
|
DAB1, reelin adaptor protein |
| chr8_+_117620317 | 10.34 |
ENSRNOT00000084220
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr7_-_134722215 | 10.26 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr14_-_83741969 | 10.22 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr11_+_45888221 | 10.13 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr2_+_187740531 | 10.11 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr4_+_13405136 | 9.97 |
ENSRNOT00000091004
|
Gnai1
|
G protein subunit alpha i1 |
| chr19_+_24329544 | 9.94 |
ENSRNOT00000080934
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr8_+_63379087 | 9.61 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr5_-_150949931 | 9.16 |
ENSRNOT00000067905
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_-_19308842 | 9.11 |
ENSRNOT00000019556
|
Fam32a
|
family with sequence similarity 32, member A |
| chr17_-_45154355 | 8.91 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr15_-_28733513 | 8.80 |
ENSRNOT00000078180
|
Sall2
|
spalt-like transcription factor 2 |
| chr8_+_116096458 | 8.20 |
ENSRNOT00000021188
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr1_+_162320730 | 8.12 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr10_-_102576878 | 8.02 |
ENSRNOT00000091129
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr17_-_90218013 | 7.68 |
ENSRNOT00000072803
|
Impad1
|
inositol monophosphatase domain containing 1 |
| chr5_+_156615886 | 7.51 |
ENSRNOT00000076439
|
Sh2d5
|
SH2 domain containing 5 |
| chrX_-_70428364 | 7.41 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr2_-_139528162 | 7.36 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr11_+_82194657 | 7.33 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr20_+_9791171 | 7.26 |
ENSRNOT00000078031
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chrX_-_71313513 | 7.09 |
ENSRNOT00000004972
ENSRNOT00000076008 ENSRNOT00000076042 |
Zmym3
|
zinc finger MYM-type containing 3 |
| chr1_+_215609036 | 6.79 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr3_-_29996865 | 6.76 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr11_+_86903122 | 6.59 |
ENSRNOT00000048063
|
Zdhhc8
|
zinc finger, DHHC-type containing 8 |
| chr1_-_198233215 | 6.54 |
ENSRNOT00000087928
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chrX_+_40258493 | 6.46 |
ENSRNOT00000033010
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr8_+_48472824 | 6.39 |
ENSRNOT00000010463
ENSRNOT00000090780 |
Mcam
|
melanoma cell adhesion molecule |
| chr3_-_125213607 | 6.03 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr9_-_81868086 | 5.74 |
ENSRNOT00000067080
|
Zfp142
|
zinc finger protein 142 |
| chrX_-_152932953 | 5.72 |
ENSRNOT00000078929
|
Cetn2
|
centrin 2 |
| chr18_+_17403407 | 5.72 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr20_+_4530342 | 5.60 |
ENSRNOT00000076352
ENSRNOT00000000478 ENSRNOT00000075925 |
Nelfe
|
negative elongation factor complex member E |
| chr10_-_70646495 | 5.38 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr11_+_83048636 | 5.36 |
ENSRNOT00000002408
|
RGD1562339
|
RGD1562339 |
| chr7_+_2605719 | 5.32 |
ENSRNOT00000018737
|
Gls2
|
glutaminase 2 |
| chr2_+_208749996 | 4.92 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr17_-_90217786 | 4.86 |
ENSRNOT00000073534
|
Impad1
|
inositol monophosphatase domain containing 1 |
| chr2_+_208750356 | 4.74 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr14_+_78973364 | 4.67 |
ENSRNOT00000089759
|
Mrfap1
|
Morf4 family associated protein 1 |
| chr4_+_145580799 | 4.60 |
ENSRNOT00000013727
|
Vhl
|
von Hippel-Lindau tumor suppressor |
| chr4_+_157511642 | 4.57 |
ENSRNOT00000065846
|
Pianp
|
PILR alpha associated neural protein |
| chr11_+_60613882 | 4.54 |
ENSRNOT00000002853
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr5_-_17663500 | 4.26 |
ENSRNOT00000033151
ENSRNOT00000034495 |
Impad1
|
inositol monophosphatase domain containing 1 |
| chr7_-_116284587 | 4.23 |
ENSRNOT00000073179
|
LOC100910656
|
ly-6/neurotoxin-like protein 1-like |
| chrX_+_22313028 | 4.21 |
ENSRNOT00000082725
|
Kdm5c
|
lysine demethylase 5C |
| chr1_-_172943853 | 4.15 |
ENSRNOT00000047040
|
Olr278
|
olfactory receptor 278 |
| chr6_-_33738825 | 4.11 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr16_-_71319449 | 4.09 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr2_+_189714754 | 4.05 |
ENSRNOT00000086285
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr3_+_152402520 | 4.00 |
ENSRNOT00000027086
|
Cnbd2
|
cyclic nucleotide binding domain containing 2 |
| chr9_-_37231291 | 3.96 |
ENSRNOT00000016237
|
Ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr12_-_16126953 | 3.95 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_+_45078556 | 3.83 |
ENSRNOT00000088280
|
Zfp192
|
zinc finger protein 192 |
| chr7_+_116980076 | 3.68 |
ENSRNOT00000012321
|
Zfp623
|
zinc finger protein 623 |
| chr1_+_220137257 | 3.56 |
ENSRNOT00000026832
|
Zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr4_+_170347410 | 3.48 |
ENSRNOT00000040508
|
LOC500350
|
LRRGT00139 |
| chr4_+_123801174 | 3.43 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr10_-_63491713 | 3.41 |
ENSRNOT00000063941
|
Tusc5
|
tumor suppressor candidate 5 |
| chr1_-_163129641 | 3.32 |
ENSRNOT00000083055
|
Capn5
|
calpain 5 |
| chr7_+_143882000 | 3.32 |
ENSRNOT00000017110
ENSRNOT00000091053 |
Mfsd5
|
major facilitator superfamily domain containing 5 |
| chr7_-_12762341 | 3.29 |
ENSRNOT00000064886
|
Abca7
|
ATP binding cassette subfamily A member 7 |
| chr1_+_255479261 | 3.20 |
ENSRNOT00000079808
|
Tnks2
|
tankyrase 2 |
| chr10_+_78168715 | 3.18 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr1_-_169721758 | 3.09 |
ENSRNOT00000023213
|
LOC100910107
|
ubiquitin carboxyl-terminal hydrolase DUB-1-like |
| chr1_-_103380338 | 3.09 |
ENSRNOT00000019061
|
Mrgprx4
|
MAS related GPR family member X4 |
| chr11_+_64761146 | 3.07 |
ENSRNOT00000051428
|
Poglut1
|
protein O-glucosyltransferase 1 |
| chr1_-_241046249 | 3.02 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr7_-_140172448 | 2.96 |
ENSRNOT00000077672
|
LOC103692976
|
cyclin-T1-like |
| chr5_-_6186329 | 2.94 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr8_+_5522739 | 2.94 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr16_-_71319052 | 2.87 |
ENSRNOT00000050980
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr2_+_211078334 | 2.86 |
ENSRNOT00000049127
|
Sort1
|
sortilin 1 |
| chr3_-_175479034 | 2.75 |
ENSRNOT00000086761
|
Hrh3
|
histamine receptor H3 |
| chr2_+_202200797 | 2.68 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr6_-_144123596 | 2.64 |
ENSRNOT00000006144
ENSRNOT00000087845 |
Wdr60
|
WD repeat domain 60 |
| chr15_-_82482009 | 2.62 |
ENSRNOT00000011926
|
Dach1
|
dachshund family transcription factor 1 |
| chr3_-_67668772 | 2.53 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr1_+_170242846 | 2.47 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr7_-_134602239 | 2.46 |
ENSRNOT00000006780
|
Zcrb1
|
zinc finger CCHC-type and RNA binding motif containing 1 |
| chr1_+_257517242 | 2.45 |
ENSRNOT00000073502
ENSRNOT00000018668 ENSRNOT00000078738 ENSRNOT00000091663 ENSRNOT00000072002 |
LOC685933
|
hypothetical protein LOC685933 |
| chr14_-_93042182 | 2.43 |
ENSRNOT00000005432
|
LOC100361025
|
protein arginine methyltransferase 1-like |
| chr8_+_70447040 | 2.43 |
ENSRNOT00000016798
|
Ints14
|
integrator complex subunit 14 |
| chr3_-_164366684 | 2.41 |
ENSRNOT00000035048
|
LOC679539
|
similar to ubiquitin-conjugating enzyme E2 variant 1 |
| chr10_+_17542374 | 2.33 |
ENSRNOT00000064079
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr10_-_49196177 | 2.27 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr10_+_34185898 | 2.24 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr1_+_172283255 | 2.24 |
ENSRNOT00000049260
|
Olr247
|
olfactory receptor 247 |
| chr3_-_119075606 | 2.20 |
ENSRNOT00000041569
ENSRNOT00000086724 |
Hdc
|
histidine decarboxylase |
| chr3_+_147894558 | 2.10 |
ENSRNOT00000055415
|
LOC502684
|
hypothetical protein LOC502684 |
| chr9_+_81868265 | 2.09 |
ENSRNOT00000022632
|
Bcs1l
|
BCS1 homolog, ubiquinol-cytochrome c reductase complex chaperone |
| chr10_+_47785033 | 1.90 |
ENSRNOT00000075073
ENSRNOT00000084944 |
B9d1
|
B9 domain containing 1 |
| chr5_-_151029233 | 1.88 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr1_-_82088275 | 1.75 |
ENSRNOT00000072616
|
Dedd2
|
death effector domain containing 2 |
| chr1_+_266451021 | 1.70 |
ENSRNOT00000027196
|
Borcs7
|
BLOC-1 related complex subunit 7 |
| chr13_+_26172243 | 1.69 |
ENSRNOT00000003840
|
Phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr10_-_84847857 | 1.63 |
ENSRNOT00000071495
|
LOC102552549
|
migration and invasion enhancer 1-like |
| chr8_-_94368834 | 1.58 |
ENSRNOT00000078977
|
Me1
|
malic enzyme 1 |
| chr1_-_219412816 | 1.56 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr1_+_172892134 | 1.47 |
ENSRNOT00000087918
|
Olr276
|
olfactory receptor 276 |
| chr1_-_172395872 | 1.38 |
ENSRNOT00000055174
|
Olr252
|
olfactory receptor 252 |
| chr1_-_90151405 | 1.38 |
ENSRNOT00000028688
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr4_+_87827582 | 1.35 |
ENSRNOT00000077211
|
LOC108348088
|
putative vomeronasal receptor-like protein 4 |
| chr20_+_3823042 | 1.25 |
ENSRNOT00000041613
|
Rxrb
|
retinoid X receptor beta |
| chr1_+_172807389 | 1.22 |
ENSRNOT00000088537
|
Olr271
|
olfactory receptor 271 |
| chr20_-_3822754 | 1.22 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr8_+_114916122 | 1.21 |
ENSRNOT00000074194
|
Tlr9
|
toll-like receptor 9 |
| chr8_-_63350269 | 1.16 |
ENSRNOT00000042681
|
Cd276
|
Cd276 molecule |
| chr1_-_44474674 | 1.10 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr10_+_94988362 | 1.04 |
ENSRNOT00000066525
|
Cep95
|
centrosomal protein 95 |
| chr1_+_213194827 | 1.01 |
ENSRNOT00000044878
|
Olr306
|
olfactory receptor 306 |
| chr13_+_37400476 | 0.94 |
ENSRNOT00000003435
|
Ccdc93
|
coiled-coil domain containing 93 |
| chr14_+_85142279 | 0.93 |
ENSRNOT00000011281
|
Thoc5
|
THO complex 5 |
| chr19_+_53629779 | 0.89 |
ENSRNOT00000051352
|
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr10_+_14689094 | 0.89 |
ENSRNOT00000036630
|
RGD1559662
|
similar to implantation serine proteinase 2 |
| chr1_+_172792874 | 0.79 |
ENSRNOT00000077837
|
Olr270
|
olfactory receptor 270 |
| chr10_-_86554478 | 0.79 |
ENSRNOT00000049621
|
Ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr10_-_82197520 | 0.78 |
ENSRNOT00000092024
|
Cacna1g
|
calcium voltage-gated channel subunit alpha1 G |
| chr16_+_7435634 | 0.75 |
ENSRNOT00000026323
|
Capn7
|
calpain 7 |
| chr6_+_93740586 | 0.65 |
ENSRNOT00000011466
|
Dact1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr1_-_172369515 | 0.63 |
ENSRNOT00000040363
|
Olr251
|
olfactory receptor 251 |
| chr2_-_177924970 | 0.58 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr7_-_120919261 | 0.57 |
ENSRNOT00000019612
|
Josd1
|
Josephin domain containing 1 |
| chr15_+_33555640 | 0.52 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr18_-_48720472 | 0.50 |
ENSRNOT00000023236
|
Cep120
|
centrosomal protein 120 |
| chr12_-_11783232 | 0.44 |
ENSRNOT00000075888
|
Zfp498
|
zinc finger protein 498 |
| chr19_+_37668693 | 0.42 |
ENSRNOT00000084970
|
Pard6a
|
par-6 family cell polarity regulator alpha |
| chr10_+_61071278 | 0.39 |
ENSRNOT00000073002
|
Olr1516
|
olfactory receptor 1516 |
| chr1_-_172932507 | 0.34 |
ENSRNOT00000048478
|
RGD1564933
|
similar to olfactory receptor Olr271 |
| chr9_-_99680979 | 0.33 |
ENSRNOT00000049347
|
Olr1345
|
olfactory receptor 1345 |
| chr10_+_14722756 | 0.31 |
ENSRNOT00000025220
|
Tpsb2
|
tryptase beta 2 |
| chr5_-_135994848 | 0.31 |
ENSRNOT00000067675
|
Btbd19
|
BTB domain containing 19 |
| chr10_+_63677396 | 0.28 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr12_+_11433813 | 0.22 |
ENSRNOT00000031901
ENSRNOT00000084577 |
Smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr16_+_80826681 | 0.21 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chr10_-_50574539 | 0.19 |
ENSRNOT00000034261
|
Cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr3_-_73636542 | 0.15 |
ENSRNOT00000074870
|
LOC100909831
|
olfactory receptor 8K3-like |
| chr1_-_216274930 | 0.07 |
ENSRNOT00000040850
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr1_+_64480929 | 0.04 |
ENSRNOT00000087016
|
Olr1l
|
olfactory receptor 1-like |
| chr9_+_100885051 | 0.04 |
ENSRNOT00000082987
|
AC109427.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.3 | 49.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 8.1 | 24.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 7.5 | 45.2 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 5.9 | 23.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 5.8 | 17.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 5.2 | 15.7 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 5.0 | 20.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 4.5 | 13.6 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 4.2 | 25.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 4.1 | 12.3 | GO:0033123 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 3.5 | 10.4 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 3.1 | 43.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.7 | 53.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 2.5 | 14.9 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 2.4 | 9.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 2.3 | 32.5 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 2.3 | 6.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 2.2 | 6.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 2.1 | 25.2 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 2.1 | 16.8 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 2.1 | 26.9 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 2.1 | 10.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 1.9 | 9.7 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.8 | 7.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 1.8 | 5.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.7 | 12.2 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 1.7 | 17.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.5 | 4.6 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 1.5 | 15.0 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 1.5 | 7.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.3 | 9.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.2 | 7.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 1.1 | 10.3 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 1.1 | 14.8 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 1.1 | 10.8 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.1 | 14.0 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.0 | 3.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 1.0 | 12.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 1.0 | 16.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 1.0 | 3.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.0 | 2.9 | GO:0060686 | esophagus smooth muscle contraction(GO:0014846) negative regulation of prostatic bud formation(GO:0060686) |
| 0.8 | 2.5 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.8 | 11.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.8 | 3.3 | GO:1902995 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.8 | 24.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.7 | 2.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.7 | 3.3 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.6 | 3.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.6 | 5.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.6 | 3.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.6 | 10.2 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.6 | 2.9 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.6 | 1.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.6 | 2.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.5 | 6.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 | 16.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 3.0 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.5 | 6.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 22.2 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.4 | 2.6 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 | 7.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.4 | 4.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 22.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.4 | 2.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.4 | 1.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) |
| 0.4 | 1.2 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.3 | 22.4 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.3 | 8.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.3 | 14.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 0.8 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.3 | 1.5 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.2 | 2.9 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 14.9 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.2 | 6.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 6.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.2 | 12.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 0.8 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.2 | 4.4 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.2 | 0.9 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 1.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.2 | 3.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.2 | 21.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 1.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.2 | 43.0 | GO:0072659 | protein localization to plasma membrane(GO:0072659) |
| 0.1 | 6.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.8 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 15.0 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 27.6 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.4 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 2.3 | GO:0042753 | negative regulation of NF-kappaB import into nucleus(GO:0042347) positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 2.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.5 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 5.7 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 8.6 | GO:0021915 | neural tube development(GO:0021915) |
| 0.1 | 1.3 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.8 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.9 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 1.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 6.1 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 2.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.9 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 2.5 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 45.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 5.5 | 16.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.9 | 49.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.9 | 5.7 | GO:0071942 | XPC complex(GO:0071942) |
| 1.8 | 42.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.5 | 10.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.4 | 5.6 | GO:0032021 | NELF complex(GO:0032021) |
| 1.4 | 24.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.1 | 36.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.7 | 27.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.7 | 2.6 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.6 | 3.0 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 6.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 2.7 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.5 | 23.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 4.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 14.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 29.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 20.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.3 | 14.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 3.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 16.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 1.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 2.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 6.5 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 18.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 10.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 3.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 9.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 25.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 2.9 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 25.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 38.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 16.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 2.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 5.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 50.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 3.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 7.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 7.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 9.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 27.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 11.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 8.1 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 16.7 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.5 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) cytoplasmic vesicle part(GO:0044433) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 42.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 6.3 | 25.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 6.1 | 49.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 5.6 | 16.8 | GO:0008254 | 3'-nucleotidase activity(GO:0008254) |
| 4.5 | 13.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 4.4 | 26.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 4.1 | 16.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 4.1 | 24.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 3.6 | 25.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 3.4 | 10.2 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 3.0 | 14.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 2.9 | 17.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 2.9 | 2.9 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 2.5 | 7.4 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 2.5 | 7.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 2.4 | 7.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 2.3 | 22.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 2.0 | 6.0 | GO:2001070 | starch binding(GO:2001070) |
| 2.0 | 24.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 1.9 | 9.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.8 | 5.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.7 | 22.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 1.7 | 10.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 1.5 | 15.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.4 | 20.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 1.4 | 14.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 1.3 | 9.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.1 | 5.7 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 1.1 | 5.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.0 | 16.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.0 | 3.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.0 | 6.8 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.9 | 6.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 12.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.9 | 7.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.7 | 2.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 11.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.5 | 3.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.5 | 10.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 1.6 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.5 | 50.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.5 | 23.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.5 | 1.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 10.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 2.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.4 | 14.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 2.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.4 | 40.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 1.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 2.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 10.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 29.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 16.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 1.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 1.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 4.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 1.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 33.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 3.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 4.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 6.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 21.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.8 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 6.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 4.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 9.3 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 17.4 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.1 | 48.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 14.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 3.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 2.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 15.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 4.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 24.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.7 | 10.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 24.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.4 | 15.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 14.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 10.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 29.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 4.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 7.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 7.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 12.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 4.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 14.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 49.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 2.5 | 10.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 2.5 | 42.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.6 | 20.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 1.0 | 25.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 1.0 | 15.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.8 | 45.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.7 | 16.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.7 | 17.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.7 | 15.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.7 | 7.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.6 | 10.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 7.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.4 | 5.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 6.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.4 | 14.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 32.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 10.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 4.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 1.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 3.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 5.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 19.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.1 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.1 | 6.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 2.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 2.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 5.0 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |