Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zbtb49
Z-value: 0.42
Transcription factors associated with Zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
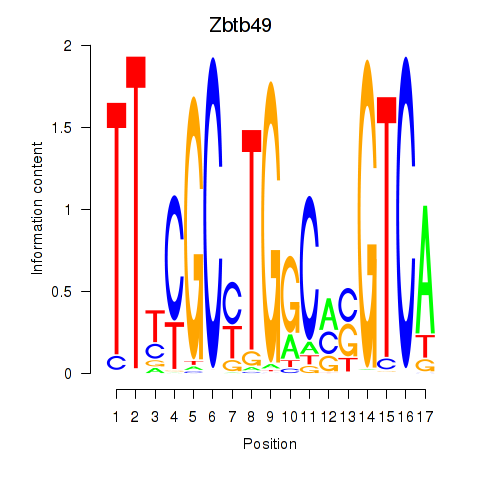
Zbtb49
|
ENSRNOG00000005633 | zinc finger and BTB domain containing 49 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb49 | rn6_v1_chr14_+_77322012_77322040 | -0.37 | 1.5e-11 | Click! |
Activity profile of Zbtb49 motif
Sorted Z-values of Zbtb49 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_248723397 | 36.32 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr1_-_258877045 | 28.54 |
ENSRNOT00000071633
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr19_+_15081590 | 23.70 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr15_-_50425900 | 12.53 |
ENSRNOT00000058715
|
Adam28
|
ADAM metallopeptidase domain 28 |
| chr7_-_70452675 | 10.26 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr20_+_2515520 | 9.99 |
ENSRNOT00000001024
|
Rpp21
|
ribonuclease P/MRP 21 subunit |
| chr15_+_32828165 | 6.61 |
ENSRNOT00000060253
|
AABR07017902.1
|
|
| chr7_+_70452579 | 6.40 |
ENSRNOT00000046099
|
B4galnt1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr9_+_14951047 | 5.97 |
ENSRNOT00000074267
|
LOC100911489
|
uncharacterized LOC100911489 |
| chr9_+_16139101 | 4.97 |
ENSRNOT00000070803
|
LOC100910668
|
uncharacterized LOC100910668 |
| chr6_+_128750795 | 4.78 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr8_+_14060394 | 4.19 |
ENSRNOT00000014827
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr4_+_149957206 | 4.07 |
ENSRNOT00000083843
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr8_+_55279373 | 3.87 |
ENSRNOT00000064290
|
Ppp2r1b
|
protein phosphatase 2 scaffold subunit A beta |
| chr10_-_13580821 | 2.94 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr6_-_128894761 | 2.80 |
ENSRNOT00000029346
|
Tcl1a
|
T-cell leukemia/lymphoma 1A |
| chr14_+_2100106 | 2.75 |
ENSRNOT00000000064
|
Gak
|
cyclin G associated kinase |
| chr8_-_22150005 | 2.64 |
ENSRNOT00000041678
|
Tyk2
|
tyrosine kinase 2 |
| chr1_+_100755682 | 2.63 |
ENSRNOT00000035748
|
Vrk3
|
vaccinia related kinase 3 |
| chr1_-_169456098 | 2.40 |
ENSRNOT00000030827
|
Trim30c
|
tripartite motif-containing 30C |
| chr7_-_143863186 | 2.11 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr8_+_116730641 | 1.95 |
ENSRNOT00000052289
|
Traip
|
TRAF-interacting protein |
| chr1_+_220746387 | 1.35 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr16_+_21178138 | 1.05 |
ENSRNOT00000027943
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr8_+_114986326 | 1.02 |
ENSRNOT00000038166
|
Poc1a
|
POC1 centriolar protein A |
| chr6_-_75589257 | 0.54 |
ENSRNOT00000006030
|
Eapp
|
E2F-associated phosphoprotein |
| chr8_+_20054422 | 0.26 |
ENSRNOT00000049132
|
Olr1162
|
olfactory receptor 1162 |
| chr9_+_49647257 | 0.16 |
ENSRNOT00000021899
|
Mrps9
|
mitochondrial ribosomal protein S9 |
| chr1_+_72272032 | 0.15 |
ENSRNOT00000050233
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb49
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 36.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.6 | 4.8 | GO:0009249 | protein lipoylation(GO:0009249) |
| 1.3 | 6.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.1 | 28.5 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.9 | 2.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.7 | 2.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.5 | 1.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 3.9 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 2.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 2.6 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 2.8 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 2.6 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 2.9 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 36.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 23.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.8 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 4.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 25.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 28.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.5 | 2.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 6.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 4.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 2.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 23.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 4.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 12.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 12.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 4.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |