Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Zbtb4
Z-value: 0.84
Transcription factors associated with Zbtb4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb4
|
ENSRNOG00000014689 | zinc finger and BTB domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb4 | rn6_v1_chr10_+_56381813_56381813 | -0.47 | 3.7e-19 | Click! |
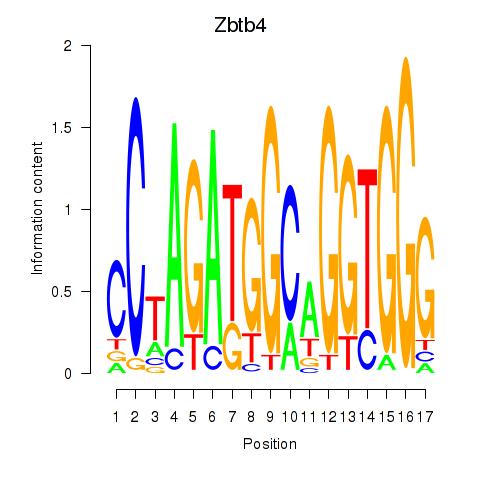
Activity profile of Zbtb4 motif
Sorted Z-values of Zbtb4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_263803150 | 55.99 |
ENSRNOT00000017840
|
Cyp2c23
|
cytochrome P450, family 2, subfamily c, polypeptide 23 |
| chr17_-_417480 | 55.00 |
ENSRNOT00000023685
|
Fbp1
|
fructose-bisphosphatase 1 |
| chrX_+_143097525 | 42.98 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr13_+_78805347 | 36.80 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr16_-_7007287 | 35.32 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_-_7007051 | 32.97 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr9_+_80118029 | 29.20 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr7_-_2677199 | 28.82 |
ENSRNOT00000043908
|
Apon
|
apolipoprotein N |
| chr12_+_52452273 | 28.36 |
ENSRNOT00000056680
ENSRNOT00000088381 |
Pxmp2
|
peroxisomal membrane protein 2 |
| chr7_-_119623072 | 26.21 |
ENSRNOT00000050511
|
Tst
|
thiosulfate sulfurtransferase |
| chr2_-_187622373 | 23.36 |
ENSRNOT00000026396
|
Rhbg
|
Rh family B glycoprotein |
| chr18_-_51651267 | 22.04 |
ENSRNOT00000020325
|
Aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr7_-_12246729 | 21.57 |
ENSRNOT00000044030
|
Reep6
|
receptor accessory protein 6 |
| chr5_+_79179417 | 21.11 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr2_-_210738378 | 20.79 |
ENSRNOT00000025746
|
Gstm6
|
glutathione S-transferase, mu 6 |
| chr13_-_102721218 | 19.31 |
ENSRNOT00000005459
|
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr4_+_154309426 | 16.38 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chr8_-_78233430 | 15.98 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr1_-_18058055 | 15.74 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr1_-_162385575 | 14.89 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr7_-_130350570 | 14.84 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr14_+_43810119 | 14.53 |
ENSRNOT00000003327
|
Rbm47
|
RNA binding motif protein 47 |
| chr5_-_164971903 | 13.94 |
ENSRNOT00000067059
|
Fbxo44
|
F-box protein 44 |
| chr1_-_101741441 | 13.43 |
ENSRNOT00000028570
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr8_+_81766041 | 12.25 |
ENSRNOT00000032280
|
Onecut1
|
one cut homeobox 1 |
| chr3_-_63211842 | 12.20 |
ENSRNOT00000008371
ENSRNOT00000050355 |
Pde11a
|
phosphodiesterase 11A |
| chr4_+_154423209 | 12.12 |
ENSRNOT00000075799
|
LOC100911545
|
alpha-2-macroglobulin-like |
| chr7_-_3342491 | 11.91 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr3_-_151106557 | 11.17 |
ENSRNOT00000025657
|
Gss
|
glutathione synthetase |
| chr1_+_143036218 | 10.94 |
ENSRNOT00000025797
|
Pde8a
|
phosphodiesterase 8A |
| chr9_-_65879521 | 10.94 |
ENSRNOT00000017517
|
Als2cr11
|
amyotrophic lateral sclerosis 2 chromosome region, candidate 11 |
| chr7_-_130347587 | 10.88 |
ENSRNOT00000040541
|
Tymp
|
thymidine phosphorylase |
| chr12_+_12227010 | 10.46 |
ENSRNOT00000060843
ENSRNOT00000092610 |
Baiap2l1
|
BAI1-associated protein 2-like 1 |
| chr8_+_117068582 | 10.41 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr7_+_12247498 | 10.10 |
ENSRNOT00000022358
|
Pcsk4
|
proprotein convertase subtilisin/kexin type 4 |
| chr7_-_124999137 | 9.72 |
ENSRNOT00000039228
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr15_-_42751330 | 9.71 |
ENSRNOT00000066478
|
Adam2
|
ADAM metallopeptidase domain 2 |
| chr1_-_80783898 | 9.48 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr4_+_22082194 | 9.41 |
ENSRNOT00000091799
|
Crot
|
carnitine O-octanoyltransferase |
| chr15_-_45927804 | 9.27 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr3_+_171743929 | 9.07 |
ENSRNOT00000079113
ENSRNOT00000064400 |
Rab22a
|
RAB22A, member RAS oncogene family |
| chr4_-_80367606 | 8.97 |
ENSRNOT00000014331
|
LOC500124
|
similar to RIKEN cDNA 4921507P07 |
| chr17_-_90756048 | 8.71 |
ENSRNOT00000085669
|
Ero1b
|
endoplasmic reticulum oxidoreductase 1 beta |
| chr6_-_135829953 | 8.48 |
ENSRNOT00000080623
ENSRNOT00000039059 |
Cdc42bpb
|
CDC42 binding protein kinase beta |
| chr10_+_56824505 | 8.33 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr1_+_153589471 | 8.27 |
ENSRNOT00000023205
|
Fzd4
|
frizzled class receptor 4 |
| chr10_+_56445647 | 7.94 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr1_-_56982554 | 7.37 |
ENSRNOT00000047157
ENSRNOT00000078312 |
Tcte3
|
t-complex-associated testis expressed 3 |
| chr20_+_46250363 | 7.21 |
ENSRNOT00000076522
ENSRNOT00000000334 |
Cd164
|
CD164 molecule |
| chr3_-_15433252 | 7.20 |
ENSRNOT00000008817
|
Lhx6
|
LIM homeobox 6 |
| chr3_-_52212412 | 7.06 |
ENSRNOT00000007615
|
Galnt3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr10_-_83822996 | 6.92 |
ENSRNOT00000008139
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr15_-_88036354 | 6.91 |
ENSRNOT00000014747
|
Ednrb
|
endothelin receptor type B |
| chr5_-_147412705 | 6.80 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chrX_+_20351486 | 6.80 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chrX_+_114929029 | 6.72 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr1_-_259106948 | 6.46 |
ENSRNOT00000074429
|
LOC100912537
|
tectonic-3-like |
| chr14_+_60857989 | 6.45 |
ENSRNOT00000034411
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr1_+_21748261 | 6.27 |
ENSRNOT00000019519
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr19_-_869490 | 6.26 |
ENSRNOT00000080433
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr5_+_159845774 | 6.24 |
ENSRNOT00000012328
|
Epha2
|
Eph receptor A2 |
| chr2_-_195888216 | 6.24 |
ENSRNOT00000056436
|
Tuft1
|
tuftelin 1 |
| chr2_-_119537837 | 6.18 |
ENSRNOT00000015200
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr3_+_55369214 | 6.10 |
ENSRNOT00000067161
|
Nostrin
|
nitric oxide synthase trafficking |
| chr5_-_136721379 | 6.04 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr12_-_44962656 | 6.01 |
ENSRNOT00000001499
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr7_+_13062196 | 5.97 |
ENSRNOT00000000193
|
Plpp2
|
phospholipid phosphatase 2 |
| chr5_-_50193571 | 5.58 |
ENSRNOT00000051243
|
Cfap206
|
cilia and flagella associated protein 206 |
| chr1_-_53038229 | 5.51 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr11_+_61661724 | 5.48 |
ENSRNOT00000079423
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr1_-_259691742 | 5.45 |
ENSRNOT00000088065
|
Tctn3
|
tectonic family member 3 |
| chr1_-_216703534 | 5.39 |
ENSRNOT00000027972
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chrX_+_32495809 | 5.26 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr8_+_13796021 | 5.15 |
ENSRNOT00000013927
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr17_-_9441526 | 5.06 |
ENSRNOT00000000146
|
Txndc15
|
thioredoxin domain containing 15 |
| chr15_+_61069581 | 5.02 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_+_87220618 | 5.01 |
ENSRNOT00000041975
|
Gsc2
|
goosecoid homeobox 2 |
| chr1_-_256013495 | 5.00 |
ENSRNOT00000023342
|
Ide
|
insulin degrading enzyme |
| chr8_-_55491152 | 4.91 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chrX_+_123404518 | 4.90 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr19_-_10101451 | 4.88 |
ENSRNOT00000017629
|
Mmp15
|
matrix metallopeptidase 15 |
| chr20_+_17750744 | 4.73 |
ENSRNOT00000000745
|
RGD1559903
|
similar to RIKEN cDNA 1700049L16 |
| chr8_+_82257849 | 4.68 |
ENSRNOT00000011963
ENSRNOT00000075708 |
Gnb5
|
G protein subunit beta 5 |
| chrX_+_29525256 | 4.63 |
ENSRNOT00000050018
|
Rab9a
|
RAB9A, member RAS oncogene family |
| chr18_-_27725694 | 4.55 |
ENSRNOT00000026398
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr8_-_6203515 | 4.53 |
ENSRNOT00000087278
ENSRNOT00000031189 ENSRNOT00000008074 ENSRNOT00000085285 ENSRNOT00000007866 |
Yap1
|
yes-associated protein 1 |
| chr5_+_58151985 | 4.47 |
ENSRNOT00000020885
|
Il11ra1
|
interleukin 11 receptor subunit alpha 1 |
| chr15_-_52332097 | 4.46 |
ENSRNOT00000017723
|
Fgf17
|
fibroblast growth factor 17 |
| chr10_-_45851984 | 4.36 |
ENSRNOT00000058308
|
Zfp496
|
zinc finger protein 496 |
| chr7_+_10962330 | 4.32 |
ENSRNOT00000008360
|
Tle6
|
transducin-like enhancer of split 6 |
| chr1_+_85496937 | 4.27 |
ENSRNOT00000076809
ENSRNOT00000087175 |
Selenov
|
selenoprotein V |
| chr5_+_152559577 | 4.27 |
ENSRNOT00000082245
|
Slc30a2
|
solute carrier family 30 member 2 |
| chr1_-_83977236 | 4.24 |
ENSRNOT00000028434
|
Egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr10_+_49299125 | 4.22 |
ENSRNOT00000074853
|
Tvp23b
|
trans-golgi network vesicle protein 23 homolog B |
| chr5_+_143500441 | 4.22 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr1_-_214074663 | 4.21 |
ENSRNOT00000026215
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr8_+_59457018 | 4.14 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr8_-_42430928 | 4.07 |
ENSRNOT00000072172
|
Olr1257
|
olfactory receptor 1257 |
| chr19_+_16417817 | 4.04 |
ENSRNOT00000015272
|
Irx5
|
iroquois homeobox 5 |
| chr11_-_45510961 | 4.02 |
ENSRNOT00000002238
|
Tomm70
|
translocase of outer mitochondrial membrane 70 |
| chr11_+_32211115 | 3.97 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr11_+_46179940 | 3.94 |
ENSRNOT00000088152
|
Tfg
|
Trk-fused gene |
| chr11_+_61661310 | 3.88 |
ENSRNOT00000093325
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr20_+_2004052 | 3.87 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr12_+_6265748 | 3.81 |
ENSRNOT00000061252
|
Wdr95
|
WD40 repeat domain 95 |
| chr7_-_120558805 | 3.76 |
ENSRNOT00000087344
|
Pla2g6
|
phospholipase A2 group VI |
| chr4_+_114935970 | 3.75 |
ENSRNOT00000082336
|
Slc4a5
|
solute carrier family 4 member 5 |
| chr16_-_12662336 | 3.75 |
ENSRNOT00000080161
|
RGD1559508
|
similar to hypothetical protein 4930474N05 |
| chr7_-_142318620 | 3.73 |
ENSRNOT00000006351
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr8_-_107380933 | 3.69 |
ENSRNOT00000022179
|
Pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr1_-_214511529 | 3.69 |
ENSRNOT00000026390
|
Chid1
|
chitinase domain containing 1 |
| chr1_-_193459396 | 3.66 |
ENSRNOT00000020281
|
Zkscan2
|
zinc finger with KRAB and SCAN domains 2 |
| chr6_-_147172022 | 3.55 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr8_-_116965396 | 3.54 |
ENSRNOT00000042528
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chr4_-_73174179 | 3.41 |
ENSRNOT00000048203
|
Tpk1
|
thiamin pyrophosphokinase 1 |
| chr9_+_47386626 | 3.41 |
ENSRNOT00000021270
|
Slc9a2
|
solute carrier family 9 member A2 |
| chr3_+_93968855 | 3.29 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chrX_+_110512013 | 3.27 |
ENSRNOT00000057044
|
Trap1a
|
tumor rejection antigen P1A |
| chr2_+_84678948 | 3.23 |
ENSRNOT00000046325
|
Fam173b
|
family with sequence similarity 173, member B |
| chr16_+_56247659 | 3.20 |
ENSRNOT00000017452
|
Tusc3
|
tumor suppressor candidate 3 |
| chr7_+_144531814 | 3.09 |
ENSRNOT00000033300
|
Hoxc13
|
homeobox C13 |
| chr20_-_45815940 | 3.05 |
ENSRNOT00000073276
|
Gpr6
|
G protein-coupled receptor 6 |
| chr18_+_31444472 | 3.01 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr10_+_20591432 | 2.97 |
ENSRNOT00000059780
|
Pank3
|
pantothenate kinase 3 |
| chr18_-_68934953 | 2.93 |
ENSRNOT00000058894
|
AABR07032503.1
|
|
| chr14_-_83741969 | 2.89 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr1_+_142136452 | 2.89 |
ENSRNOT00000016445
|
Hddc3
|
HD domain containing 3 |
| chr7_-_122967178 | 2.86 |
ENSRNOT00000052098
|
Rangap1
|
RAN GTPase activating protein 1 |
| chr1_+_240908483 | 2.75 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr1_+_48625743 | 2.72 |
ENSRNOT00000064018
ENSRNOT00000084317 |
Map3k4
|
mitogen activated protein kinase kinase kinase 4 |
| chr9_+_6966908 | 2.66 |
ENSRNOT00000072796
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr13_-_73819896 | 2.65 |
ENSRNOT00000036392
|
Fam163a
|
family with sequence similarity 163, member A |
| chr9_+_53627208 | 2.65 |
ENSRNOT00000083487
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr20_-_11340296 | 2.53 |
ENSRNOT00000038547
|
Icoslg
|
inducible T-cell co-stimulator ligand |
| chr4_-_11497531 | 2.50 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr20_+_7762327 | 2.46 |
ENSRNOT00000000595
|
Zfp523
|
zinc finger protein 523 |
| chr10_-_38969501 | 2.40 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr9_+_64095978 | 2.33 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr7_+_25832521 | 2.32 |
ENSRNOT00000084955
|
Rfx4
|
regulatory factor X4 |
| chr1_+_101704746 | 2.27 |
ENSRNOT00000028564
|
Fam83e
|
family with sequence similarity 83, member E |
| chr1_-_276012351 | 2.25 |
ENSRNOT00000045642
|
AABR07007000.1
|
|
| chr7_-_59514939 | 2.21 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr5_-_59292944 | 2.20 |
ENSRNOT00000051234
|
Olr838
|
olfactory receptor 838 |
| chr8_-_126495347 | 2.19 |
ENSRNOT00000013467
|
Cmc1
|
C-x(9)-C motif containing 1 |
| chr8_+_40505439 | 2.19 |
ENSRNOT00000076145
|
Olr1205-ps
|
olfactory receptor 1205, pseudogene |
| chr3_-_2456041 | 2.19 |
ENSRNOT00000062001
|
Rnf224
|
ring finger protein 224 |
| chr18_-_74574052 | 2.17 |
ENSRNOT00000079899
ENSRNOT00000091439 |
Slc14a2
|
solute carrier family 14 member 2 |
| chr18_+_70263359 | 2.15 |
ENSRNOT00000019573
|
Cfap53
|
cilia and flagella associated protein 53 |
| chr7_+_121480723 | 2.11 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr4_-_123161985 | 2.10 |
ENSRNOT00000011490
|
Xpc
|
XPC complex subunit, DNA damage recognition and repair factor |
| chr13_-_77959089 | 2.09 |
ENSRNOT00000003586
|
Cacybp
|
calcyclin binding protein |
| chr13_-_93746994 | 2.08 |
ENSRNOT00000005072
|
Opn3
|
opsin 3 |
| chr12_+_22026075 | 2.05 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr7_-_115979298 | 2.03 |
ENSRNOT00000075883
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr5_-_76039760 | 2.01 |
ENSRNOT00000019959
|
RGD1306148
|
similar to KIAA0368 |
| chr7_+_16363599 | 2.00 |
ENSRNOT00000044356
|
Olr1059
|
olfactory receptor 1059 |
| chr6_-_26792808 | 1.97 |
ENSRNOT00000010403
|
Abhd1
|
abhydrolase domain containing 1 |
| chrX_+_10964067 | 1.91 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
| chr10_+_60974225 | 1.91 |
ENSRNOT00000042349
|
Olr1511
|
olfactory receptor 1511 |
| chr18_+_73645907 | 1.90 |
ENSRNOT00000023487
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr4_+_158224000 | 1.88 |
ENSRNOT00000084240
ENSRNOT00000078495 |
Ano2
|
anoctamin 2 |
| chr10_-_15667955 | 1.82 |
ENSRNOT00000085746
ENSRNOT00000027940 |
Mpg
|
N-methylpurine-DNA glycosylase |
| chr8_+_40524733 | 1.78 |
ENSRNOT00000076626
ENSRNOT00000074528 |
Olr1206
|
olfactory receptor 1206 |
| chr18_+_29110242 | 1.73 |
ENSRNOT00000025756
|
Pura
|
purine rich element binding protein A |
| chr10_-_61088827 | 1.71 |
ENSRNOT00000047421
|
Olr1517
|
olfactory receptor 1517 |
| chr10_-_90049112 | 1.66 |
ENSRNOT00000028323
|
Pyy
|
peptide YY (mapped) |
| chr12_+_31335372 | 1.65 |
ENSRNOT00000001242
ENSRNOT00000037296 |
Stx2
|
syntaxin 2 |
| chr8_-_48692295 | 1.63 |
ENSRNOT00000065456
|
Vps11
|
VPS11 CORVET/HOPS core subunit |
| chr11_-_74793673 | 1.62 |
ENSRNOT00000002338
ENSRNOT00000002343 |
Opa1
|
OPA1, mitochondrial dynamin like GTPase |
| chr16_+_74292438 | 1.62 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr9_-_121972055 | 1.59 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr7_-_83701447 | 1.57 |
ENSRNOT00000088893
|
AABR07057675.1
|
|
| chr7_+_11992543 | 1.49 |
ENSRNOT00000024576
|
Abhd17a
|
abhydrolase domain containing 17A |
| chr1_+_224970807 | 1.49 |
ENSRNOT00000026141
|
Tmem223
|
transmembrane protein 223 |
| chr8_+_41898624 | 1.48 |
ENSRNOT00000073212
|
Olr1218
|
olfactory receptor 1218 |
| chr8_+_65686648 | 1.45 |
ENSRNOT00000017233
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr7_+_122700854 | 1.44 |
ENSRNOT00000086603
|
Rbx1
|
ring-box 1 |
| chr16_-_70665796 | 1.34 |
ENSRNOT00000080051
ENSRNOT00000084974 |
Thap1
|
THAP domain containing 1 |
| chr7_-_58219790 | 1.31 |
ENSRNOT00000067907
|
Tbc1d15
|
TBC1 domain family, member 15 |
| chr12_+_11347643 | 1.31 |
ENSRNOT00000038175
ENSRNOT00000075986 |
Kpna7
|
karyopherin subunit alpha 7 |
| chr12_-_52644260 | 1.28 |
ENSRNOT00000073986
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr5_-_99043546 | 1.28 |
ENSRNOT00000055741
|
Oog3
|
oogenesin 3 |
| chr16_+_21282467 | 1.26 |
ENSRNOT00000065345
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr2_+_157316266 | 1.24 |
ENSRNOT00000015387
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr8_-_39734594 | 1.21 |
ENSRNOT00000076872
|
Slc37a2
|
solute carrier family 37 member 2 |
| chr5_-_106865746 | 1.15 |
ENSRNOT00000008224
|
Ifnb1
|
interferon beta 1 |
| chr4_+_176629975 | 1.13 |
ENSRNOT00000086529
|
Spx
|
spexin hormone |
| chr7_-_12918173 | 1.12 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr10_+_60997525 | 1.12 |
ENSRNOT00000084195
|
Olr1512
|
olfactory receptor 1512 |
| chr8_-_53362006 | 1.09 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr3_+_65815080 | 1.08 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_-_59313689 | 1.06 |
ENSRNOT00000047909
|
Olr839
|
olfactory receptor 839 |
| chr7_-_11982141 | 1.02 |
ENSRNOT00000024980
|
Scamp4
|
secretory carrier membrane protein 4 |
| chr13_-_83680207 | 1.01 |
ENSRNOT00000004199
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chr11_+_77239098 | 1.00 |
ENSRNOT00000058701
|
Gmnc
|
geminin coiled-coil domain containing |
| chr10_+_36098051 | 0.96 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr9_+_10535340 | 0.96 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr4_-_1752548 | 0.95 |
ENSRNOT00000071677
|
LOC100912415
|
olfactory receptor 150-like |
| chr5_+_140803638 | 0.91 |
ENSRNOT00000050949
|
Bmp8b
|
bone morphogenetic protein 8b |
| chr13_-_110077946 | 0.90 |
ENSRNOT00000000078
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.7 | 55.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 9.2 | 36.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 8.7 | 26.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 7.8 | 23.4 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 4.8 | 19.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 4.1 | 16.4 | GO:1990402 | embryonic liver development(GO:1990402) |
| 3.6 | 14.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 2.8 | 22.0 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 2.8 | 2.8 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 2.6 | 21.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 2.6 | 10.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 2.5 | 19.7 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 2.3 | 6.8 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 2.2 | 68.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 2.1 | 56.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.9 | 9.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 1.8 | 5.5 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.7 | 8.7 | GO:0030070 | insulin processing(GO:0030070) |
| 1.7 | 6.9 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 1.7 | 5.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.6 | 6.2 | GO:1901491 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) negative regulation of lymphangiogenesis(GO:1901491) |
| 1.4 | 7.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 1.4 | 8.3 | GO:0042701 | extracellular matrix-cell signaling(GO:0035426) progesterone secretion(GO:0042701) retinal blood vessel morphogenesis(GO:0061304) |
| 1.1 | 3.4 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.1 | 15.7 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 1.1 | 21.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.1 | 2.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.0 | 27.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.9 | 3.8 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.9 | 12.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.9 | 5.4 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.9 | 6.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) positive regulation of corticotropin secretion(GO:0051461) |
| 0.9 | 6.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.9 | 11.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.8 | 7.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.8 | 2.4 | GO:1903660 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.8 | 2.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.7 | 3.7 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 3.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.7 | 4.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.7 | 2.1 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.7 | 10.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 2.7 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.6 | 4.5 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.6 | 2.5 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.6 | 3.7 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.6 | 3.6 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.6 | 3.5 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.6 | 14.9 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.5 | 6.3 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.5 | 5.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.5 | 2.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.5 | 10.9 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.4 | 0.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.4 | 1.7 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.4 | 2.9 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.4 | 10.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.4 | 4.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.4 | 9.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.4 | 4.9 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 | 4.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.4 | 4.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.4 | 10.9 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.4 | 2.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 3.1 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 3.7 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.3 | 43.0 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.3 | 2.5 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 | 8.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.3 | 9.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.3 | 3.9 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.3 | 12.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.3 | 3.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.2 | 1.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 9.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 4.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 8.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 1.2 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) |
| 0.2 | 1.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 2.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 3.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.4 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 4.5 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 3.9 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.2 | 1.7 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.1 | 0.9 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 7.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 4.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 2.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 2.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 2.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 4.9 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 3.0 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.9 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.7 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 5.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 8.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 1.9 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 0.9 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 0.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 6.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 2.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 2.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 7.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 5.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 2.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 3.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.0 | GO:0019098 | reproductive behavior(GO:0019098) |
| 0.0 | 0.7 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 2.0 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.6 | GO:0044317 | rod spherule(GO:0044317) |
| 2.3 | 23.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.8 | 3.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 1.5 | 4.5 | GO:0071149 | TEAD-1-YAP complex(GO:0071148) TEAD-2-YAP complex(GO:0071149) |
| 1.5 | 28.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 1.2 | 3.5 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.0 | 2.9 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.9 | 5.6 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.9 | 6.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.8 | 4.9 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.7 | 2.1 | GO:1990590 | Lewy body core(GO:1990037) ATF4-CREB1 transcription factor complex(GO:1990589) ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.7 | 2.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.7 | 9.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.6 | 6.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.5 | 9.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 62.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.5 | 5.0 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.4 | 1.3 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.3 | 1.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.3 | 1.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 7.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 3.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 16.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 1.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 3.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 1.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 25.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 5.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 3.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 12.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 11.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 9.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 17.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 48.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 2.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 4.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 11.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 47.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 7.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.9 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 0.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 200.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 3.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 5.1 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.1 | 1.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 5.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 5.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.7 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 5.4 | 10.9 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 4.8 | 19.3 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 4.5 | 13.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 4.1 | 16.4 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 3.9 | 55.0 | GO:0016208 | AMP binding(GO:0016208) |
| 3.2 | 29.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 3.1 | 9.4 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 2.1 | 6.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 1.9 | 56.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.7 | 12.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.4 | 22.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 1.2 | 6.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 1.1 | 4.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 1.1 | 5.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 15.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 1.0 | 4.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.0 | 117.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 1.0 | 2.9 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.9 | 4.5 | GO:0005111 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.8 | 23.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.8 | 6.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.7 | 3.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.7 | 3.7 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.7 | 2.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.6 | 7.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 5.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.6 | 2.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 7.9 | GO:0008061 | chitin binding(GO:0008061) |
| 0.6 | 10.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.6 | 4.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 2.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.6 | 12.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.6 | 1.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 6.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.6 | 6.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.5 | 8.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.5 | 6.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 11.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 20.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 4.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.5 | 3.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.5 | 10.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.5 | 3.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 8.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 9.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 3.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.4 | 3.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 1.1 | GO:0031765 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 0.3 | 3.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 6.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 2.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.3 | 3.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 3.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.3 | 9.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 7.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 4.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 2.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 0.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 43.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 11.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 1.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 0.9 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 4.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 17.0 | GO:0008144 | drug binding(GO:0008144) |
| 0.1 | 3.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 15.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.6 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 3.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.6 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 3.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 4.9 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 1.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.9 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 6.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.0 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 2.0 | GO:0016298 | lipase activity(GO:0016298) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.9 | GO:0015085 | calcium channel activity(GO:0005262) calcium ion transmembrane transporter activity(GO:0015085) |
| 0.0 | 0.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 4.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 36.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.6 | 16.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.6 | 151.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 8.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 12.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 6.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 8.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 6.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 3.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 2.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 8.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 18.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 43.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.5 | 29.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.3 | 15.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.2 | 49.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 1.0 | 13.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.0 | 26.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.9 | 10.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.9 | 16.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 7.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 14.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 9.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 11.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 12.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 4.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 6.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.3 | 5.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 2.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 6.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 4.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 6.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.7 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.2 | 4.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 4.9 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 7.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 22.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 18.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.9 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 7.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 0.9 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.1 | 0.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 3.9 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 4.0 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 4.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |