Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
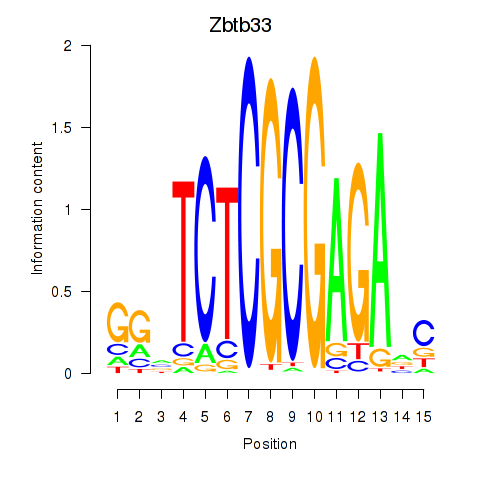
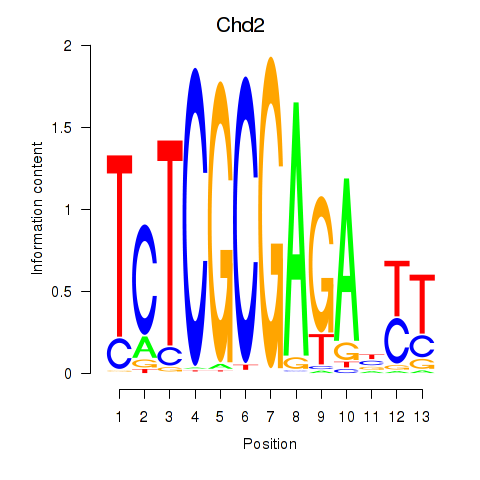
Results for Zbtb33_Chd2
Z-value: 1.84
Transcription factors associated with Zbtb33_Chd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb33
|
ENSRNOG00000028899 | zinc finger and BTB domain containing 33 |
|
Chd2
|
ENSRNOG00000012716 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Chd2 | rn6_v1_chr1_-_134867001_134867001 | 0.33 | 2.5e-09 | Click! |
| Zbtb33 | rn6_v1_chrX_+_124393332_124393332 | 0.15 | 8.9e-03 | Click! |
Activity profile of Zbtb33_Chd2 motif
Sorted Z-values of Zbtb33_Chd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_69927938 | 83.46 |
ENSRNOT00000058838
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr5_+_38844488 | 68.86 |
ENSRNOT00000009545
|
Mms22l
|
MMS22-like, DNA repair protein |
| chr10_-_19652898 | 65.91 |
ENSRNOT00000009648
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr11_-_54404612 | 49.97 |
ENSRNOT00000033837
|
RGD1310335
|
similar to RIKEN cDNA C330027C09 |
| chr2_+_165601007 | 47.27 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr7_+_140758615 | 47.19 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr1_+_279633671 | 41.14 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr9_-_11027506 | 37.72 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr3_-_110021149 | 37.08 |
ENSRNOT00000007808
|
Fsip1
|
fibrous sheath interacting protein 1 |
| chr1_+_257615088 | 36.88 |
ENSRNOT00000083183
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr5_-_137265015 | 36.40 |
ENSRNOT00000036151
|
Cdc20
|
cell division cycle 20 |
| chr6_+_144291974 | 33.74 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr1_+_257614731 | 33.13 |
ENSRNOT00000075680
|
LOC100911660
|
lymphocyte-specific helicase-like |
| chr1_+_257766691 | 32.11 |
ENSRNOT00000088148
|
Hells
|
helicase, lymphoid specific |
| chr1_+_146976975 | 31.60 |
ENSRNOT00000071895
|
LOC100366231
|
X-linked lymphocyte-regulated 5C-like |
| chr2_-_187771857 | 31.53 |
ENSRNOT00000092517
ENSRNOT00000035383 |
Pmf1
|
polyamine-modulated factor 1 |
| chr1_-_261051498 | 31.14 |
ENSRNOT00000071417
|
Arhgap19
|
Rho GTPase activating protein 19 |
| chr1_+_257901985 | 30.59 |
ENSRNOT00000073015
|
Hells
|
helicase, lymphoid specific |
| chr10_-_86932154 | 29.23 |
ENSRNOT00000085344
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr10_+_86157608 | 26.60 |
ENSRNOT00000082668
ENSRNOT00000008256 |
Cdk12
|
cyclin-dependent kinase 12 |
| chr9_+_98279022 | 25.68 |
ENSRNOT00000038536
|
Rbm44
|
RNA binding motif protein 44 |
| chr20_+_5933303 | 23.20 |
ENSRNOT00000000617
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr2_+_257425679 | 22.98 |
ENSRNOT00000066199
|
Fubp1
|
far upstream element binding protein 1 |
| chr9_-_78368777 | 22.88 |
ENSRNOT00000020414
|
Bard1
|
BRCA1 associated RING domain 1 |
| chr7_-_126028279 | 22.46 |
ENSRNOT00000044883
|
Smc1b
|
structural maintenance of chromosomes 1B |
| chr10_-_59698168 | 22.38 |
ENSRNOT00000074018
|
Gsg2
|
germ cell associated 2 (haspin) |
| chr9_+_20765296 | 22.10 |
ENSRNOT00000016291
|
Cd2ap
|
CD2-associated protein |
| chr6_+_132702448 | 21.64 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr2_-_185444897 | 21.46 |
ENSRNOT00000016329
|
Rps3a
|
ribosomal protein S3a |
| chr20_+_7908304 | 21.04 |
ENSRNOT00000000603
|
Rpl10a
|
ribosomal protein L10A |
| chr12_-_52452040 | 20.56 |
ENSRNOT00000067453
|
Pole
|
DNA polymerase epsilon, catalytic subunit |
| chr2_+_208577388 | 19.51 |
ENSRNOT00000022571
|
Wdr77
|
WD repeat domain 77 |
| chr13_+_105408179 | 19.36 |
ENSRNOT00000003378
|
Ddx3y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr10_-_18090932 | 19.29 |
ENSRNOT00000064744
ENSRNOT00000006591 |
Npm1
|
nucleophosmin 1 |
| chr2_+_208420206 | 19.13 |
ENSRNOT00000090500
ENSRNOT00000076065 ENSRNOT00000073644 ENSRNOT00000086279 |
LOC100911453
Wdr77
|
methylosome protein 50-like WD repeat domain 77 |
| chr1_-_104202591 | 19.12 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr11_+_86092468 | 19.05 |
ENSRNOT00000057971
|
LOC100361706
|
lambda-chain C1-region-like |
| chr15_-_89339323 | 18.77 |
ENSRNOT00000013297
|
Rbm26
|
RNA binding motif protein 26 |
| chr5_-_157573183 | 18.00 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr16_+_81521073 | 17.91 |
ENSRNOT00000066521
|
Upf3a
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr15_-_89339037 | 17.68 |
ENSRNOT00000013255
|
Rbm26
|
RNA binding motif protein 26 |
| chr15_-_38096875 | 17.15 |
ENSRNOT00000032723
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr2_+_208577197 | 17.00 |
ENSRNOT00000076654
|
Wdr77
|
WD repeat domain 77 |
| chr3_+_4867233 | 16.96 |
ENSRNOT00000006754
|
Rpl7a
|
ribosomal protein L7a |
| chr20_+_5933595 | 16.36 |
ENSRNOT00000000618
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr3_+_12009578 | 16.32 |
ENSRNOT00000021725
|
Rpl12
|
ribosomal protein L12 |
| chr2_-_205426869 | 16.15 |
ENSRNOT00000023333
|
Sycp1
|
synaptonemal complex protein 1 |
| chr1_+_31967978 | 16.03 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr16_+_71011332 | 15.80 |
ENSRNOT00000086891
ENSRNOT00000020181 |
Ash2l
|
ASH2 like histone lysine methyltransferase complex subunit |
| chr9_-_86129073 | 15.74 |
ENSRNOT00000093730
|
Cul3
|
cullin 3 |
| chrX_-_38196060 | 15.73 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr12_+_16410083 | 15.56 |
ENSRNOT00000001709
|
Mad1l1
|
MAD1 mitotic arrest deficient like 1 |
| chr4_+_115024927 | 15.53 |
ENSRNOT00000090987
|
Mob1a
|
MOB kinase activator 1A |
| chr3_+_46286712 | 15.50 |
ENSRNOT00000085563
|
March7
|
membrane associated ring-CH-type finger 7 |
| chr19_-_37819789 | 15.22 |
ENSRNOT00000036702
|
Cenpt
|
centromere protein T |
| chr4_+_123162086 | 15.22 |
ENSRNOT00000011557
|
Lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_138194136 | 15.18 |
ENSRNOT00000014928
|
RGD1307595
|
similar to RIKEN cDNA 1700018B24 |
| chr8_+_99977334 | 15.07 |
ENSRNOT00000085808
ENSRNOT00000056704 ENSRNOT00000041859 |
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr4_-_180887050 | 14.86 |
ENSRNOT00000065730
|
Ints13
|
integrator complex subunit 13 |
| chr6_-_47904437 | 14.81 |
ENSRNOT00000092867
|
Rps7
|
ribosomal protein S7 |
| chr1_-_89474252 | 14.78 |
ENSRNOT00000028597
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr6_-_47904163 | 14.61 |
ENSRNOT00000011333
|
Rps7
|
ribosomal protein S7 |
| chr1_-_101118825 | 14.39 |
ENSRNOT00000066328
|
Rps11
|
ribosomal protein S11 |
| chr18_+_44468784 | 13.88 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr13_+_47440803 | 13.46 |
ENSRNOT00000030476
|
Yod1
|
YOD1 deubiquitinase |
| chr2_+_187708234 | 13.46 |
ENSRNOT00000026514
|
Smg5
|
SMG5 nonsense mediated mRNA decay factor |
| chr12_+_19599834 | 13.39 |
ENSRNOT00000092039
ENSRNOT00000042006 |
Stag3
|
stromal antigen 3 |
| chr3_-_117389456 | 13.29 |
ENSRNOT00000007103
ENSRNOT00000081533 |
Myef2
|
myelin expression factor 2 |
| chr13_-_70321722 | 13.00 |
ENSRNOT00000092397
ENSRNOT00000092336 ENSRNOT00000092649 ENSRNOT00000092269 |
Smg7
|
SMG7 nonsense mediated mRNA decay factor |
| chr2_-_208152179 | 12.91 |
ENSRNOT00000068152
|
Ddx20
|
DEAD-box helicase 20 |
| chr15_+_24159647 | 12.88 |
ENSRNOT00000082675
|
Lgals3
|
galectin 3 |
| chr10_-_55997299 | 12.87 |
ENSRNOT00000074505
|
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr5_-_57845509 | 12.86 |
ENSRNOT00000035541
|
Kif24
|
kinesin family member 24 |
| chr3_-_152259156 | 12.85 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chr10_-_94460732 | 12.82 |
ENSRNOT00000014508
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr18_-_11789697 | 12.74 |
ENSRNOT00000077304
ENSRNOT00000064554 |
Dsc3
|
desmocollin 3 |
| chr1_+_248228496 | 12.65 |
ENSRNOT00000015406
|
Uhrf2
|
ubiquitin like with PHD and ring finger domains 2 |
| chr19_+_10596960 | 12.62 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr8_+_59457018 | 12.30 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr5_+_122508388 | 12.25 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr1_-_178196294 | 12.04 |
ENSRNOT00000067170
|
Btbd10
|
BTB domain containing 10 |
| chrX_+_120860178 | 12.01 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr14_-_5859581 | 11.80 |
ENSRNOT00000052308
|
AABR07014241.1
|
|
| chr7_+_18497807 | 11.44 |
ENSRNOT00000011094
|
Zfp414
|
zinc finger protein 414 |
| chr3_-_151724654 | 11.40 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr3_+_150055749 | 11.39 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr6_+_137243185 | 11.19 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr2_+_166402838 | 10.65 |
ENSRNOT00000064244
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr15_-_59215803 | 10.54 |
ENSRNOT00000032301
|
Lacc1
|
laccase domain containing 1 |
| chr10_+_93415399 | 10.48 |
ENSRNOT00000066484
|
Tlk2
|
tousled-like kinase 2 |
| chr1_-_64021321 | 10.33 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr13_-_73638073 | 10.31 |
ENSRNOT00000005195
|
Cep350
|
centrosomal protein 350 |
| chr6_+_34155017 | 10.13 |
ENSRNOT00000073663
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr10_-_86144880 | 10.01 |
ENSRNOT00000067901
|
Med1
|
mediator complex subunit 1 |
| chrX_+_120859968 | 9.95 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr12_+_37594185 | 9.82 |
ENSRNOT00000088787
|
Sbno1
|
strawberry notch homolog 1 |
| chr2_-_165600748 | 9.74 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr15_-_40545824 | 9.69 |
ENSRNOT00000017204
|
Nup58
|
nucleoporin 58 |
| chr12_+_23954574 | 9.67 |
ENSRNOT00000046343
|
Styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr9_+_65279675 | 9.65 |
ENSRNOT00000019486
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr1_-_187766670 | 9.61 |
ENSRNOT00000092513
ENSRNOT00000092282 |
Rps15a
|
ribosomal protein S15a |
| chr7_-_28715224 | 9.59 |
ENSRNOT00000065899
|
Parpbp
|
PARP1 binding protein |
| chr2_-_216509629 | 9.49 |
ENSRNOT00000023366
ENSRNOT00000087714 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr19_+_57650163 | 9.33 |
ENSRNOT00000038257
ENSRNOT00000083572 |
Sprtn
|
SprT-like N-terminal domain |
| chr14_-_20992753 | 9.30 |
ENSRNOT00000014650
|
Mob1b
|
MOB kinase activator 1B |
| chr2_-_58534211 | 9.27 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr1_+_163663407 | 9.26 |
ENSRNOT00000088873
|
Thap12
|
THAP domain containing 12 |
| chr6_-_137808303 | 9.11 |
ENSRNOT00000019689
|
Brf1
|
BRF1, RNA polymerase III transcription initiation factor 90 subunit |
| chr9_+_17120759 | 9.10 |
ENSRNOT00000025787
|
Polr1c
|
RNA polymerase I subunit C |
| chrX_+_29562165 | 9.06 |
ENSRNOT00000006074
|
Ofd1
|
OFD1, centriole and centriolar satellite protein |
| chrX_-_123731294 | 8.95 |
ENSRNOT00000092574
ENSRNOT00000032618 |
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr5_-_151117042 | 8.91 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr10_-_56185857 | 8.90 |
ENSRNOT00000014261
|
Wrap53
|
WD repeat containing, antisense to TP53 |
| chr11_-_27141881 | 8.90 |
ENSRNOT00000002169
|
Cct8
|
chaperonin containing TCP1 subunit 8 |
| chr1_-_64090017 | 8.81 |
ENSRNOT00000086622
ENSRNOT00000091654 |
Rps9
|
ribosomal protein S9 |
| chr14_-_59735450 | 8.79 |
ENSRNOT00000071929
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr8_+_72029489 | 8.75 |
ENSRNOT00000089336
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr13_+_25656983 | 8.75 |
ENSRNOT00000020892
|
RGD1307235
|
similar to RIKEN cDNA 2310035C23 |
| chr1_+_205706468 | 8.64 |
ENSRNOT00000089957
ENSRNOT00000023877 |
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr1_-_32140136 | 8.55 |
ENSRNOT00000081339
ENSRNOT00000022635 |
Slc12a7
|
solute carrier family 12 member 7 |
| chrX_-_14783792 | 8.50 |
ENSRNOT00000087609
|
LOC691895
|
similar to ferritin, heavy polypeptide-like 17 |
| chr16_+_64729221 | 8.37 |
ENSRNOT00000015297
|
Mak16
|
MAK16 homolog |
| chr16_+_81756971 | 8.26 |
ENSRNOT00000044085
|
Pcid2
|
PCI domain containing 2 |
| chr10_-_104263071 | 8.26 |
ENSRNOT00000005347
|
Grb2
|
growth factor receptor bound protein 2 |
| chr13_-_83680207 | 8.26 |
ENSRNOT00000004199
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chr4_+_45555077 | 8.20 |
ENSRNOT00000089007
|
Lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr17_-_12669573 | 8.19 |
ENSRNOT00000016942
ENSRNOT00000041726 |
Syk
|
spleen associated tyrosine kinase |
| chr5_+_113592919 | 8.17 |
ENSRNOT00000011336
|
Ift74
|
intraflagellar transport 74 |
| chr12_+_37593874 | 8.16 |
ENSRNOT00000057902
|
Sbno1
|
strawberry notch homolog 1 |
| chr10_-_108425206 | 8.13 |
ENSRNOT00000073140
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr18_-_30781292 | 8.03 |
ENSRNOT00000027125
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr17_-_955615 | 7.96 |
ENSRNOT00000022884
|
Fancc
|
Fanconi anemia, complementation group C |
| chr10_-_88355678 | 7.88 |
ENSRNOT00000076625
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr1_-_56982554 | 7.84 |
ENSRNOT00000047157
ENSRNOT00000078312 |
Tcte3
|
t-complex-associated testis expressed 3 |
| chr14_+_35683442 | 7.82 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr19_+_14523554 | 7.78 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr8_+_107826424 | 7.67 |
ENSRNOT00000019769
|
Dbr1
|
debranching RNA lariats 1 |
| chr2_-_216509827 | 7.64 |
ENSRNOT00000088533
|
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr1_-_90246265 | 7.56 |
ENSRNOT00000078585
ENSRNOT00000030186 |
Lsm14a
|
LSM14A mRNA processing body assembly factor |
| chr13_-_89874008 | 7.52 |
ENSRNOT00000051368
|
Ptma
|
prothymosin alpha |
| chr10_-_15245757 | 7.49 |
ENSRNOT00000060156
|
Wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr1_-_178195948 | 7.49 |
ENSRNOT00000081767
|
Btbd10
|
BTB domain containing 10 |
| chr2_-_28423762 | 7.41 |
ENSRNOT00000022864
|
Btf3
|
basic transcription factor 3 |
| chr10_-_95407997 | 7.40 |
ENSRNOT00000077888
|
Nol11
|
nucleolar protein 11 |
| chr4_+_118167294 | 7.39 |
ENSRNOT00000022367
|
LOC687679
|
similar to small nuclear ribonucleoprotein polypeptide G |
| chr15_+_30621374 | 7.23 |
ENSRNOT00000090632
|
AABR07017748.2
|
|
| chr1_-_274107138 | 7.11 |
ENSRNOT00000078670
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr6_+_22302069 | 7.11 |
ENSRNOT00000039132
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_-_241046249 | 7.07 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr5_+_151796814 | 7.05 |
ENSRNOT00000009394
|
Gpatch3
|
G patch domain containing 3 |
| chr17_+_78915604 | 7.04 |
ENSRNOT00000057855
|
Rpp38
|
ribonuclease P/MRP 38 subunit |
| chr8_-_58253688 | 7.03 |
ENSRNOT00000010956
|
Cul5
|
cullin 5 |
| chr7_-_18683440 | 6.96 |
ENSRNOT00000068323
|
Rps28
|
ribosomal protein S28 |
| chr12_+_24367199 | 6.92 |
ENSRNOT00000001971
|
Fkbp6
|
FK506 binding protein 6 |
| chr7_-_62162453 | 6.68 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr1_+_174132798 | 6.61 |
ENSRNOT00000019247
|
Rpl27a
|
ribosomal protein L27a |
| chr1_+_176854816 | 6.60 |
ENSRNOT00000087461
|
Usp47
|
ubiquitin specific peptidase 47 |
| chr12_-_38093050 | 6.52 |
ENSRNOT00000057814
|
Ccdc62
|
coiled-coil domain containing 62 |
| chr16_+_71090045 | 6.52 |
ENSRNOT00000020832
|
Ddhd2
|
DDHD domain containing 2 |
| chrX_-_156155014 | 6.46 |
ENSRNOT00000088637
|
LOC102552182
|
L antigen family member 3-like |
| chr10_-_95212111 | 6.44 |
ENSRNOT00000020795
|
Kpna2
|
karyopherin subunit alpha 2 |
| chrX_+_15171499 | 6.43 |
ENSRNOT00000008399
|
Suv39h1l1
|
suppressor of variegation 3-9 homolog 1 (Drosophila)-like 1 |
| chrX_+_71681264 | 6.42 |
ENSRNOT00000036989
|
LOC102553785
|
uncharacterized LOC102553785 |
| chr10_-_104155717 | 6.35 |
ENSRNOT00000089894
|
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr1_-_229639187 | 6.31 |
ENSRNOT00000016804
|
Zfp91
|
zinc finger protein 91 |
| chr4_+_34282625 | 6.30 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr1_+_114186853 | 6.23 |
ENSRNOT00000055989
|
Tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr10_+_55492404 | 6.22 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr14_-_20953095 | 6.11 |
ENSRNOT00000004440
|
Dck
|
deoxycytidine kinase |
| chr1_+_259926537 | 6.10 |
ENSRNOT00000073537
|
NEWGENE_1306399
|
cyclin J |
| chr3_+_138398011 | 6.10 |
ENSRNOT00000038865
|
Mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr20_+_4572100 | 6.10 |
ENSRNOT00000000476
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr19_-_52206310 | 6.00 |
ENSRNOT00000087886
|
Mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr17_-_70325855 | 6.00 |
ENSRNOT00000024952
|
Gdi2
|
GDP dissociation inhibitor 2 |
| chr3_-_177201525 | 5.99 |
ENSRNOT00000022451
|
Rgs19
|
regulator of G-protein signaling 19 |
| chr10_-_73690860 | 5.85 |
ENSRNOT00000004876
ENSRNOT00000075843 |
Ints2
|
integrator complex subunit 2 |
| chr15_+_31469953 | 5.82 |
ENSRNOT00000077849
|
AABR07017825.4
|
|
| chr5_-_139853247 | 5.71 |
ENSRNOT00000056644
|
Exo5
|
exonuclease 5 |
| chr16_+_21132147 | 5.65 |
ENSRNOT00000084243
ENSRNOT00000027846 |
Mau2
|
MAU2 sister chromatid cohesion factor |
| chr6_+_26517840 | 5.62 |
ENSRNOT00000031842
|
Ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr4_+_180887182 | 5.60 |
ENSRNOT00000042270
ENSRNOT00000002476 |
Fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr2_+_196402837 | 5.56 |
ENSRNOT00000086849
|
Cdc42se1
|
CDC42 small effector 1 |
| chr20_-_21689553 | 5.54 |
ENSRNOT00000038095
|
Tmem26
|
transmembrane protein 26 |
| chr3_+_65815080 | 5.45 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr8_+_69121682 | 5.41 |
ENSRNOT00000013461
|
Rpl4
|
ribosomal protein L4 |
| chr17_+_27496353 | 5.38 |
ENSRNOT00000077994
ENSRNOT00000019040 ENSRNOT00000089736 |
Ssr1
|
signal sequence receptor subunit 1 |
| chr3_-_117766120 | 5.27 |
ENSRNOT00000056022
|
Fbn1
|
fibrillin 1 |
| chrY_-_1238420 | 5.23 |
ENSRNOT00000092078
|
Ddx3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3 |
| chr6_+_99817431 | 5.19 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr10_-_90356242 | 5.18 |
ENSRNOT00000028496
|
Slc25a39
|
solute carrier family 25, member 39 |
| chr3_-_138397870 | 5.17 |
ENSRNOT00000074606
|
Snx5
|
sorting nexin 5 |
| chr1_-_156296161 | 4.96 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr10_-_65443981 | 4.95 |
ENSRNOT00000035657
|
Rpl23a
|
ribosomal protein L23a |
| chr5_+_47186558 | 4.88 |
ENSRNOT00000007654
|
LOC100910771
|
mitogen-activated protein kinase kinase kinase 7-like |
| chrX_+_156227830 | 4.88 |
ENSRNOT00000083949
|
LOC690348
|
similar to ESO3 protein |
| chr2_+_120266933 | 4.83 |
ENSRNOT00000015192
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr18_+_1142782 | 4.82 |
ENSRNOT00000046718
|
Thoc1
|
THO complex 1 |
| chr18_-_6587080 | 4.81 |
ENSRNOT00000040815
|
LOC103694404
|
60S ribosomal protein L39 |
| chr1_+_176854653 | 4.80 |
ENSRNOT00000065712
|
Usp47
|
ubiquitin specific peptidase 47 |
| chr3_+_33641616 | 4.79 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb33_Chd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 62.7 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 9.9 | 39.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 9.6 | 125.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 9.3 | 55.6 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 8.1 | 8.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 6.9 | 103.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 6.4 | 19.3 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 5.5 | 22.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 5.4 | 21.6 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 5.2 | 36.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 5.1 | 20.6 | GO:0006272 | leading strand elongation(GO:0006272) |
| 5.0 | 15.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 4.9 | 29.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 4.8 | 19.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 4.5 | 13.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 4.3 | 12.9 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 3.9 | 15.7 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 3.4 | 17.0 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 3.2 | 35.6 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 3.2 | 19.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) chorionic trophoblast cell differentiation(GO:0060718) |
| 3.0 | 9.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 2.9 | 8.8 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 2.8 | 8.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 2.7 | 8.2 | GO:0032752 | serotonin secretion by platelet(GO:0002554) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) beta selection(GO:0043366) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 2.7 | 16.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 2.5 | 10.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 2.4 | 21.5 | GO:1904851 | positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 2.3 | 16.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 2.3 | 22.9 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 2.2 | 6.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 2.1 | 72.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 2.1 | 10.6 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 2.1 | 12.7 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 2.1 | 14.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 1.9 | 3.9 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 1.9 | 14.9 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 1.8 | 33.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.8 | 5.3 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 1.6 | 15.7 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 1.6 | 15.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 1.5 | 4.5 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 1.4 | 51.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 1.4 | 37.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.4 | 4.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 1.4 | 4.2 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 1.4 | 4.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 1.4 | 24.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 1.3 | 9.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.3 | 3.9 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 1.3 | 3.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.2 | 3.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 1.2 | 3.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 1.2 | 7.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 1.2 | 8.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.2 | 2.3 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 1.2 | 8.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.2 | 3.5 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 1.1 | 5.7 | GO:0034088 | maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 1.1 | 12.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 1.1 | 4.3 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 1.1 | 28.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 1.1 | 13.9 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 1.1 | 9.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 1.0 | 6.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 1.0 | 22.8 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 1.0 | 6.1 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.0 | 8.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.0 | 12.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 1.0 | 5.8 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.9 | 4.7 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.9 | 39.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.9 | 8.3 | GO:0003360 | brainstem development(GO:0003360) |
| 0.9 | 6.4 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.9 | 3.5 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.9 | 2.6 | GO:0072098 | embryonic skeletal limb joint morphogenesis(GO:0036023) intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.9 | 3.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.8 | 0.8 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.8 | 22.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.8 | 23.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.8 | 50.0 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.8 | 7.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.8 | 7.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.8 | 9.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.8 | 1.5 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.7 | 8.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.7 | 3.7 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.7 | 9.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.7 | 15.9 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.7 | 7.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.7 | 6.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.7 | 2.8 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.7 | 15.5 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.7 | 19.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.7 | 8.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.7 | 8.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.7 | 4.0 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.7 | 6.6 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.6 | 4.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.6 | 4.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.6 | 3.5 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.6 | 2.2 | GO:0070370 | cellular heat acclimation(GO:0070370) |
| 0.6 | 6.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.5 | 2.7 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.5 | 15.2 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.5 | 2.6 | GO:2000584 | mesenchymal cell proliferation involved in lung development(GO:0060916) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.5 | 5.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 9.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.5 | 7.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.5 | 8.9 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.5 | 8.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.5 | 2.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.5 | 5.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.5 | 13.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 11.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.5 | 1.4 | GO:0015817 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.4 | 8.0 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.4 | 6.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 6.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.4 | 2.1 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.4 | 1.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.4 | 10.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.4 | 0.8 | GO:0060526 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.4 | 2.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 4.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 30.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.4 | 2.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 5.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.3 | 15.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.3 | 14.6 | GO:0007129 | synapsis(GO:0007129) |
| 0.3 | 2.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.3 | 5.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.3 | 3.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.3 | 5.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 4.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 12.3 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.3 | 2.5 | GO:0034650 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.3 | 9.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.3 | 0.5 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.3 | 1.3 | GO:0033483 | gas homeostasis(GO:0033483) |
| 0.3 | 4.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 15.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 1.0 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 9.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 7.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 0.7 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.2 | 12.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 34.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 4.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 3.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 2.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 20.1 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.2 | 4.3 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 1.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 11.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.2 | 0.7 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 | 6.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 1.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 4.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 6.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 3.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 7.7 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.1 | 11.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.1 | 2.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 5.0 | GO:0045814 | negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.1 | 1.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 1.6 | GO:2000269 | regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 | 1.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 3.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 3.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 6.0 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.7 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 9.3 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 1.2 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.0 | 9.6 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 15.8 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 1.0 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 2.3 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 3.6 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 6.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 4.2 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
| 0.0 | 5.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 3.1 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 7.7 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 9.1 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 9.0 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.0 | GO:0051884 | anagen(GO:0042640) regulation of anagen(GO:0051884) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.8 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.0 | 68.9 | GO:0035101 | FACT complex(GO:0035101) |
| 20.6 | 164.5 | GO:0000796 | condensin complex(GO:0000796) |
| 12.6 | 37.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 9.8 | 29.5 | GO:0000802 | transverse filament(GO:0000802) |
| 8.9 | 26.6 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 7.9 | 31.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 7.3 | 29.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 5.9 | 83.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 5.7 | 22.9 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 4.4 | 4.4 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 4.1 | 20.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 3.7 | 22.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 3.7 | 55.6 | GO:0034709 | methylosome(GO:0034709) |
| 3.0 | 15.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 2.8 | 19.3 | GO:0001652 | granular component(GO:0001652) |
| 2.5 | 7.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 2.2 | 15.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 2.2 | 8.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 2.1 | 6.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 2.1 | 8.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 2.0 | 8.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 2.0 | 8.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 2.0 | 40.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 2.0 | 62.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 1.8 | 12.9 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.8 | 22.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 1.7 | 17.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.6 | 119.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.5 | 40.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 1.5 | 7.4 | GO:0034455 | t-UTP complex(GO:0034455) |
| 1.4 | 7.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.4 | 35.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 1.3 | 4.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 1.3 | 11.7 | GO:0042555 | MCM complex(GO:0042555) |
| 1.3 | 6.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.3 | 21.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 1.2 | 7.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.2 | 4.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.2 | 7.0 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 1.2 | 22.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.0 | 10.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.0 | 9.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 1.0 | 12.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 1.0 | 12.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.9 | 4.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.9 | 4.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.9 | 3.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.9 | 7.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.9 | 5.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.8 | 15.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.8 | 9.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.7 | 0.7 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.7 | 60.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 36.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.6 | 4.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 29.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.6 | 26.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.6 | 8.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.6 | 15.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.6 | 12.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 14.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.5 | 8.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 3.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 5.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 3.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.5 | 11.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.4 | 49.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.4 | 12.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.4 | 8.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 5.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 12.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.3 | 5.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 4.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 9.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 20.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 7.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 13.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 2.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 8.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 28.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 4.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 19.3 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 5.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 2.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 8.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 2.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 2.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 21.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 2.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 2.1 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.2 | 25.7 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.2 | 3.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.0 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 4.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 1.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 4.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 6.8 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 3.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 26.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 10.5 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 10.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 22.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 7.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 2.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.2 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.1 | 4.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 4.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 65.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 7.3 | 29.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 7.3 | 36.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 5.7 | 17.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 5.2 | 15.7 | GO:0031208 | POZ domain binding(GO:0031208) |
| 5.0 | 15.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 4.9 | 39.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) NFAT protein binding(GO:0051525) |
| 4.7 | 37.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 4.3 | 21.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 4.1 | 20.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 3.9 | 11.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 3.8 | 19.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 3.7 | 29.4 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 3.1 | 12.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 3.1 | 12.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 2.8 | 22.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 2.7 | 8.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.6 | 55.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 2.6 | 12.9 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 2.3 | 49.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 2.3 | 25.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 2.3 | 9.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 1.8 | 22.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 1.8 | 27.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.7 | 17.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.7 | 35.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 1.6 | 8.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 1.5 | 31.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 1.5 | 6.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) |
| 1.4 | 4.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 1.3 | 21.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 1.2 | 31.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 1.1 | 4.5 | GO:0035877 | death effector domain binding(GO:0035877) |
| 1.1 | 17.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 1.0 | 9.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 1.0 | 10.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.9 | 3.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.9 | 8.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.9 | 22.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.9 | 4.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.8 | 12.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.8 | 2.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.8 | 3.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.7 | 6.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.7 | 8.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.7 | 7.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.7 | 2.8 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.7 | 4.8 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.7 | 5.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 5.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.6 | 44.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.6 | 33.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.6 | 3.8 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.6 | 7.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 23.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.6 | 59.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.6 | 6.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 6.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.5 | 6.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 16.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 12.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.5 | 3.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.5 | 9.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.5 | 2.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 2.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.5 | 18.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.5 | 1.4 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.4 | 3.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 2.9 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) cyclohydrolase activity(GO:0019238) |
| 0.4 | 12.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 5.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 2.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 4.2 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.4 | 6.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 9.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 6.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 8.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 8.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.3 | 2.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 9.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 38.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 3.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 14.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.7 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 4.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 11.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 6.2 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.2 | 8.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 6.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 2.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 48.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 8.2 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.2 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 2.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 9.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 4.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 9.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 3.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 4.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 3.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 4.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 14.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 5.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 7.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 9.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 18.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 4.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 3.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 4.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.0 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 1.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 14.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.3 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.1 | 5.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 35.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 107.4 | GO:0005524 | ATP binding(GO:0005524) |
| 0.1 | 2.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 2.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 7.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 5.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 4.2 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 4.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 5.1 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 1.4 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 22.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 3.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 6.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 18.0 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 164.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 2.3 | 47.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 1.3 | 95.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 1.2 | 83.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 1.0 | 34.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.7 | 30.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.7 | 32.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.5 | 19.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.5 | 13.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.4 | 12.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 9.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 6.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 6.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.3 | 3.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 4.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.3 | 21.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 9.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 8.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 5.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 16.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 6.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 6.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 9.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 3.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 11.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 39.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 2.5 | 40.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 2.1 | 117.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.8 | 3.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 1.5 | 136.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 1.5 | 25.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 1.4 | 68.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 1.2 | 8.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 1.1 | 24.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 1.1 | 11.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.0 | 4.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 1.0 | 15.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.0 | 70.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.9 | 12.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.8 | 23.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.8 | 22.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.8 | 6.2 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.8 | 70.7 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.7 | 8.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.7 | 18.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.6 | 5.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.5 | 6.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.5 | 1.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.4 | 10.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 4.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.4 | 6.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.4 | 12.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 8.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.4 | 8.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 8.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 10.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.3 | 5.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 3.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 9.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 15.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 2.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 3.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 3.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 9.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 4.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 24.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 16.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 2.3 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 2.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.4 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 2.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 5.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |