Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
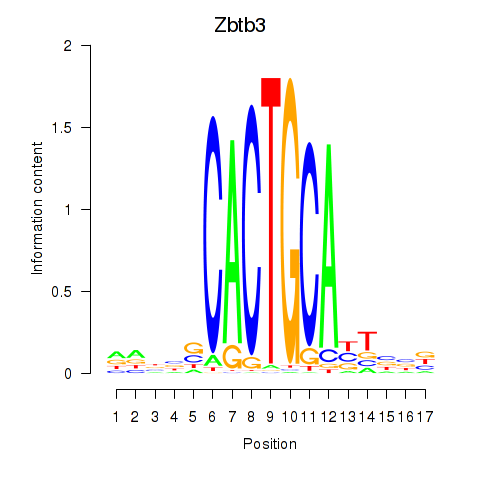
Results for Zbtb3
Z-value: 1.07
Transcription factors associated with Zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb3
|
ENSRNOG00000019461 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb3 | rn6_v1_chr1_+_224998172_224998172 | 0.25 | 5.9e-06 | Click! |
Activity profile of Zbtb3 motif
Sorted Z-values of Zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_84939323 | 107.43 |
ENSRNOT00000022742
|
Myo16
|
myosin XVI |
| chr7_-_113937941 | 59.93 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chrX_+_65040934 | 48.69 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr1_-_188713270 | 48.52 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr3_-_176666282 | 48.28 |
ENSRNOT00000016947
|
Eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr16_+_26906716 | 47.42 |
ENSRNOT00000064297
|
Cpe
|
carboxypeptidase E |
| chrX_+_65040775 | 45.61 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr2_+_18392142 | 44.82 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr20_-_13142856 | 41.91 |
ENSRNOT00000001743
|
S100b
|
S100 calcium binding protein B |
| chr2_-_26699333 | 41.39 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr2_-_147959567 | 39.04 |
ENSRNOT00000063986
|
LOC100909840
|
profilin-2-like |
| chr3_+_71210301 | 38.00 |
ENSRNOT00000006504
|
Fam171b
|
family with sequence similarity 171, member B |
| chr1_+_80195532 | 37.21 |
ENSRNOT00000022528
|
Rtn2
|
reticulon 2 |
| chr18_+_30435119 | 34.47 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr18_-_37096132 | 32.77 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr2_+_220298245 | 32.55 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr8_-_73194837 | 31.30 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr6_+_29977797 | 31.18 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr1_-_89560719 | 29.46 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr2_-_148050423 | 27.99 |
ENSRNOT00000064506
ENSRNOT00000023469 |
LOC100909840
|
profilin-2-like |
| chr2_-_210943620 | 27.63 |
ENSRNOT00000026750
|
Gpr61
|
G protein-coupled receptor 61 |
| chr1_+_226435979 | 27.53 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr15_+_5306459 | 27.15 |
ENSRNOT00000092079
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr11_+_69739384 | 24.98 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_+_157726941 | 24.39 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr12_-_5682608 | 24.26 |
ENSRNOT00000076483
|
Fry
|
FRY microtubule binding protein |
| chr3_+_48082935 | 23.28 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr7_-_25743537 | 22.57 |
ENSRNOT00000083450
|
LOC100910996
|
uncharacterized LOC100910996 |
| chr2_+_218951451 | 21.10 |
ENSRNOT00000019190
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr6_+_119519714 | 20.60 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_+_15966234 | 19.83 |
ENSRNOT00000084304
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr5_+_139963002 | 18.09 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr12_-_6740714 | 18.04 |
ENSRNOT00000001205
|
Medag
|
mesenteric estrogen-dependent adipogenesis |
| chr1_+_101427195 | 17.87 |
ENSRNOT00000028271
|
Gys1
|
glycogen synthase 1 |
| chr1_+_48077033 | 17.77 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr3_+_142739781 | 17.77 |
ENSRNOT00000006181
|
Sstr4
|
somatostatin receptor 4 |
| chr7_+_139762614 | 17.54 |
ENSRNOT00000031157
|
Ccdc184
|
coiled-coil domain containing 184 |
| chrX_+_75792931 | 17.25 |
ENSRNOT00000037575
|
Magee2
|
MAGE family member E2 |
| chr20_+_28572242 | 16.99 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr15_-_60752106 | 16.96 |
ENSRNOT00000058148
|
Akap11
|
A-kinase anchoring protein 11 |
| chr3_+_172692452 | 16.31 |
ENSRNOT00000077193
|
Zfp831
|
zinc finger protein 831 |
| chr13_-_90641772 | 15.68 |
ENSRNOT00000064601
|
Atp1a4
|
ATPase Na+/K+ transporting subunit alpha 4 |
| chr3_+_161433410 | 15.60 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr1_+_16478127 | 15.23 |
ENSRNOT00000019076
|
Ahi1
|
Abelson helper integration site 1 |
| chr15_+_41448064 | 15.11 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr1_-_261333383 | 15.05 |
ENSRNOT00000037490
|
Morn4
|
MORN repeat containing 4 |
| chr14_+_79540235 | 14.31 |
ENSRNOT00000089334
|
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr2_+_218951141 | 14.14 |
ENSRNOT00000091001
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chrX_+_92131209 | 14.09 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr1_+_67340809 | 13.11 |
ENSRNOT00000045266
|
LOC100362054
|
mCG114696-like |
| chr7_+_143749221 | 12.29 |
ENSRNOT00000014807
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr2_+_44096061 | 11.54 |
ENSRNOT00000018438
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr12_-_15996958 | 10.87 |
ENSRNOT00000039726
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr20_+_13067242 | 10.75 |
ENSRNOT00000083878
|
Dip2a
|
disco-interacting protein 2 homolog A |
| chr3_+_149802621 | 10.53 |
ENSRNOT00000071772
|
LOC690507
|
similar to Vomeromodulin |
| chr15_+_43582020 | 10.40 |
ENSRNOT00000013001
|
Pnma2
|
paraneoplastic Ma antigen 2 |
| chr1_-_124039196 | 10.32 |
ENSRNOT00000051883
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr4_+_70903253 | 10.21 |
ENSRNOT00000019720
|
Ephb6
|
Eph receptor B6 |
| chr7_+_122979021 | 10.16 |
ENSRNOT00000085707
|
Zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr1_+_273854248 | 9.44 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr17_-_88037034 | 9.39 |
ENSRNOT00000080193
ENSRNOT00000034907 |
Prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr18_-_29340403 | 8.93 |
ENSRNOT00000025157
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr2_+_180012414 | 8.76 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chr7_+_18510354 | 8.66 |
ENSRNOT00000060002
|
Pram1
|
PML-RARA regulated adaptor molecule 1 |
| chr12_-_47919400 | 8.54 |
ENSRNOT00000073774
|
Mvk
|
mevalonate kinase |
| chr4_-_77706994 | 8.39 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr6_+_137967401 | 8.34 |
ENSRNOT00000029223
|
LOC690422
|
hypothetical protein LOC690422 |
| chr4_-_98141482 | 8.27 |
ENSRNOT00000009883
|
Il12rb2
|
interleukin 12 receptor subunit beta 2 |
| chr1_+_80982358 | 8.21 |
ENSRNOT00000078462
|
AABR07002683.1
|
|
| chr15_+_39945095 | 7.92 |
ENSRNOT00000016826
|
Shisa2
|
shisa family member 2 |
| chr3_-_76496283 | 7.74 |
ENSRNOT00000044305
|
Olr611
|
olfactory receptor 611 |
| chrX_-_17252877 | 7.49 |
ENSRNOT00000060213
|
LOC102547118
|
retinitis pigmentosa 1-like 1 protein-like |
| chr2_+_72006099 | 7.44 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr17_-_34945317 | 7.20 |
ENSRNOT00000090457
|
Vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_+_201256910 | 7.19 |
ENSRNOT00000090143
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_60088406 | 6.87 |
ENSRNOT00000091192
|
Vom1r7
|
vomeronasal 1 receptor 7 |
| chr3_-_44522930 | 6.85 |
ENSRNOT00000006963
|
Acvr1
|
activin A receptor type 1 |
| chr3_+_67849966 | 6.82 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr10_+_103934797 | 6.82 |
ENSRNOT00000035865
|
Cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr5_-_150723321 | 6.39 |
ENSRNOT00000018038
|
Atpif1
|
ATPase inhibitory factor 1 |
| chr18_+_87580424 | 6.25 |
ENSRNOT00000070984
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr1_-_172322795 | 6.23 |
ENSRNOT00000075318
|
RGD1562400
|
similar to olfactory receptor MOR204-14 |
| chr5_+_128450680 | 6.14 |
ENSRNOT00000010700
|
Txndc12
|
thioredoxin domain containing 12 |
| chr2_-_28799266 | 5.55 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr1_+_201055644 | 5.45 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr4_+_2711385 | 5.21 |
ENSRNOT00000035996
|
Dnajb6
|
DnaJ heat shock protein family (Hsp40) member B6 |
| chr9_+_16702460 | 5.14 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr19_-_10358695 | 4.98 |
ENSRNOT00000019770
|
Katnb1
|
katanin regulatory subunit B1 |
| chr11_-_71128299 | 4.76 |
ENSRNOT00000080568
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr1_-_261446570 | 4.75 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr11_-_67702955 | 4.49 |
ENSRNOT00000084540
ENSRNOT00000078285 |
Kpna1
|
karyopherin subunit alpha 1 |
| chr18_-_56870801 | 4.47 |
ENSRNOT00000087188
ENSRNOT00000032189 |
Arhgef37
|
Rho guanine nucleotide exchange factor 37 |
| chr8_+_44990014 | 4.25 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr19_+_561727 | 4.13 |
ENSRNOT00000016259
ENSRNOT00000081547 |
Rrad
|
RRAD, Ras related glycolysis inhibitor and calcium channel regulator |
| chr3_-_29996865 | 4.04 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_1410339 | 4.00 |
ENSRNOT00000035905
|
Heatr5b
|
HEAT repeat containing 5B |
| chr1_+_172807389 | 3.92 |
ENSRNOT00000088537
|
Olr271
|
olfactory receptor 271 |
| chr7_-_120636563 | 3.80 |
ENSRNOT00000091317
ENSRNOT00000031117 |
Tmem184b
|
transmembrane protein 184B |
| chr8_-_128622314 | 3.79 |
ENSRNOT00000084950
ENSRNOT00000024408 |
Gorasp1
|
golgi reassembly stacking protein 1 |
| chr5_+_173340060 | 3.77 |
ENSRNOT00000067841
|
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr1_+_35067490 | 3.75 |
ENSRNOT00000022790
|
Adamts16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr3_-_120373500 | 3.74 |
ENSRNOT00000067727
|
Nphp1
|
nephrocystin 1 |
| chr14_+_86652365 | 3.60 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr15_-_3773340 | 3.54 |
ENSRNOT00000063831
|
LOC103689945
|
zinc finger SWIM domain-containing protein 8-like |
| chr2_-_86475096 | 3.39 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr18_+_60639860 | 3.24 |
ENSRNOT00000083363
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_+_45966926 | 3.23 |
ENSRNOT00000067584
|
Olfr225
|
olfactory receptor 225 |
| chr19_+_9587653 | 3.22 |
ENSRNOT00000015956
|
Got2
|
glutamic-oxaloacetic transaminase 2 |
| chr4_-_176026133 | 3.20 |
ENSRNOT00000043374
ENSRNOT00000046598 |
Slco1a4
|
solute carrier organic anion transporter family, member 1a4 |
| chr4_+_113247795 | 3.16 |
ENSRNOT00000007984
|
Tacr1
|
tachykinin receptor 1 |
| chrX_-_124576133 | 3.14 |
ENSRNOT00000041904
|
Rhox3
|
reproductive homeobox on X chromosome 3 |
| chr1_-_168082740 | 3.13 |
ENSRNOT00000070789
|
LOC684376
|
similar to olfactory receptor 555 |
| chr4_-_146954159 | 3.05 |
ENSRNOT00000010397
|
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr8_+_31497639 | 2.87 |
ENSRNOT00000020041
|
Snx19
|
sorting nexin 19 |
| chr2_-_211235440 | 2.85 |
ENSRNOT00000033015
ENSRNOT00000091896 |
Sars
|
seryl-tRNA synthetase |
| chr1_+_266451021 | 2.67 |
ENSRNOT00000027196
|
Borcs7
|
BLOC-1 related complex subunit 7 |
| chr18_-_16543992 | 2.66 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr2_+_186980793 | 2.53 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr2_-_144217600 | 2.51 |
ENSRNOT00000000101
|
Rfxapl1
|
regulatory factor X-associated protein-like 1 |
| chr10_+_35343189 | 2.43 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr7_-_18180705 | 2.42 |
ENSRNOT00000065954
|
Zfp494
|
zinc finger protein 494 |
| chr5_+_173340487 | 2.36 |
ENSRNOT00000078306
|
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr5_-_62683964 | 2.35 |
ENSRNOT00000060005
|
Anks6
|
ankyrin repeat and sterile alpha motif domain containing 6 |
| chr4_-_100303047 | 2.34 |
ENSRNOT00000018170
ENSRNOT00000084782 |
Mat2a
|
methionine adenosyltransferase 2A |
| chr2_-_144007636 | 2.24 |
ENSRNOT00000092028
|
Rfxap
|
regulatory factor X-associated protein |
| chr5_+_69214948 | 2.14 |
ENSRNOT00000074807
|
Olr844
|
olfactory receptor 844 |
| chr6_+_1410507 | 2.11 |
ENSRNOT00000034919
|
Gpatch11
|
G patch domain containing 11 |
| chr8_-_22929294 | 1.97 |
ENSRNOT00000015609
|
Rab3d
|
RAB3D, member RAS oncogene family |
| chr6_+_64252020 | 1.95 |
ENSRNOT00000047296
ENSRNOT00000082105 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr3_+_150188455 | 1.91 |
ENSRNOT00000072917
|
AABR07054370.1
|
|
| chr14_-_81426496 | 1.80 |
ENSRNOT00000018073
|
Add1
|
adducin 1 |
| chr10_-_12818137 | 1.77 |
ENSRNOT00000060972
|
Olr1382
|
olfactory receptor 1382 |
| chr2_-_210991509 | 1.72 |
ENSRNOT00000087329
|
Cyb561d1
|
cytochrome b561 family, member D1 |
| chr1_+_265883355 | 1.31 |
ENSRNOT00000025938
|
Elovl3
|
ELOVL fatty acid elongase 3 |
| chr5_-_131860637 | 1.25 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr1_-_101426852 | 1.18 |
ENSRNOT00000028217
|
Ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr1_+_172792874 | 1.17 |
ENSRNOT00000077837
|
Olr270
|
olfactory receptor 270 |
| chr16_+_3817402 | 1.16 |
ENSRNOT00000015011
|
Tmem254
|
transmembrane protein 254 |
| chr15_+_36565495 | 1.07 |
ENSRNOT00000028093
|
Atp12a
|
ATPase H+/K+ transporting non-gastric alpha2 subunit |
| chr9_+_93799278 | 1.01 |
ENSRNOT00000084590
|
Dis3l2
|
DIS3-like 3'-5' exoribonuclease 2 |
| chr7_-_122963299 | 1.00 |
ENSRNOT00000090208
|
Rangap1
|
RAN GTPase activating protein 1 |
| chr15_+_31533675 | 0.99 |
ENSRNOT00000084141
|
AABR07017825.7
|
|
| chr9_+_100885051 | 0.95 |
ENSRNOT00000082987
|
AC109427.1
|
|
| chr5_-_166430254 | 0.79 |
ENSRNOT00000048914
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr5_+_166430400 | 0.70 |
ENSRNOT00000021822
|
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr10_+_13915214 | 0.69 |
ENSRNOT00000015092
|
Pkd1
|
polycystin 1, transient receptor potential channel interacting |
| chr16_+_82184387 | 0.62 |
ENSRNOT00000089329
ENSRNOT00000068416 |
Tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chrX_-_75566481 | 0.59 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr7_-_11783849 | 0.51 |
ENSRNOT00000026202
|
Sf3a2
|
splicing factor 3a, subunit 2 |
| chr1_-_204817080 | 0.51 |
ENSRNOT00000077956
|
Fam53b
|
family with sequence similarity 53, member B |
| chr4_-_51003117 | 0.36 |
ENSRNOT00000034936
|
Tas2r118
|
taste receptor, type 2, member 118 |
| chr13_+_49850189 | 0.25 |
ENSRNOT00000042125
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr3_+_73366155 | 0.04 |
ENSRNOT00000044979
|
Olr473
|
olfactory receptor 473 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 9.5 | 47.4 | GO:2000173 | insulin processing(GO:0030070) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 9.2 | 27.5 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 6.1 | 24.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 6.0 | 41.9 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 5.9 | 29.5 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 5.6 | 67.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 5.0 | 25.0 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 4.6 | 27.5 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 4.5 | 31.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 3.8 | 15.2 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 3.7 | 48.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 3.7 | 7.4 | GO:0050893 | sensory processing(GO:0050893) |
| 3.1 | 9.4 | GO:0006178 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 3.0 | 17.8 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 2.8 | 8.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 2.8 | 31.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 2.6 | 10.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 2.4 | 4.7 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 2.2 | 8.9 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 2.2 | 32.8 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 1.8 | 20.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.7 | 15.6 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.7 | 5.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.6 | 6.4 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 1.5 | 107.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 1.5 | 23.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 1.4 | 8.7 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 1.4 | 4.3 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 1.4 | 17.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 1.3 | 4.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.2 | 6.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 1.2 | 3.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 1.1 | 23.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 1.1 | 3.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 1.1 | 3.2 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 1.0 | 16.7 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.9 | 19.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.9 | 12.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.8 | 32.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.8 | 3.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.7 | 48.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.6 | 37.2 | GO:0065002 | intracellular protein transmembrane transport(GO:0065002) |
| 0.6 | 1.2 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.6 | 7.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 17.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.5 | 7.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.5 | 2.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.5 | 12.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.5 | 1.9 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.5 | 2.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.5 | 5.1 | GO:0060484 | establishment of epithelial cell apical/basal polarity(GO:0045198) lung-associated mesenchyme development(GO:0060484) |
| 0.4 | 3.0 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.4 | 4.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 6.1 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.4 | 8.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 2.4 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 1.8 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 18.0 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.3 | 8.3 | GO:0045622 | regulation of T-helper cell differentiation(GO:0045622) |
| 0.3 | 41.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 0.8 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.3 | 1.3 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 4.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 2.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.7 | GO:0072021 | ascending thin limb development(GO:0072021) distal tubule morphogenesis(GO:0072156) metanephric ascending thin limb development(GO:0072218) metanephric proximal tubule development(GO:0072237) |
| 0.2 | 3.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 3.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 41.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.2 | 1.0 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 7.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.0 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.1 | 27.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.6 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 3.7 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.1 | 42.4 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.1 | 0.6 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.5 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.0 | 1.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 4.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 4.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 2.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 13.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.1 | 48.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 8.1 | 24.4 | GO:0035577 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 6.0 | 18.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 4.7 | 75.0 | GO:0031045 | dense core granule(GO:0031045) |
| 3.9 | 23.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.8 | 12.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.6 | 31.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.6 | 107.4 | GO:0016459 | myosin complex(GO:0016459) |
| 1.5 | 29.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.4 | 4.3 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 1.4 | 6.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 1.3 | 15.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.2 | 37.2 | GO:0098827 | endoplasmic reticulum subcompartment(GO:0098827) |
| 1.2 | 32.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.0 | 31.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.9 | 15.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.8 | 2.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 7.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.5 | 59.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.5 | 67.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.5 | 41.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.4 | 15.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 1.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.3 | 7.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.2 | 6.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 41.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 0.6 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 2.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 15.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 1.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 29.2 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 3.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 34.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 20.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.0 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 6.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 15.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 54.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 4.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 3.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 5.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 52.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 8.7 | GO:0030054 | cell junction(GO:0030054) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 35.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 6.7 | 67.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 5.9 | 29.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 4.7 | 41.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 4.0 | 47.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 3.0 | 17.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 3.0 | 32.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 2.8 | 31.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 2.6 | 17.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 2.5 | 17.8 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 2.3 | 6.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 2.2 | 48.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 2.1 | 6.4 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.6 | 44.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.6 | 6.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 1.5 | 20.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.4 | 12.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.3 | 15.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.3 | 5.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.2 | 16.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.2 | 16.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.1 | 6.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 1.1 | 25.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.1 | 3.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 1.1 | 4.3 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 1.1 | 3.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 1.0 | 3.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.9 | 8.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.8 | 23.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.8 | 15.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.8 | 2.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.8 | 107.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.7 | 17.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.6 | 7.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.6 | 43.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.6 | 24.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.6 | 59.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.5 | 3.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 10.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.5 | 32.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 6.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.4 | 27.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.4 | 8.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 5.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 31.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 5.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 0.8 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.2 | 1.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 2.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.2 | 8.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 9.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 4.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 4.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 3.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 2.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 13.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 12.6 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 6.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 12.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 4.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 37.7 | GO:0046872 | metal ion binding(GO:0046872) |
| 0.0 | 4.3 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 25.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.9 | 47.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.8 | 34.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.7 | 32.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.7 | 17.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.5 | 6.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.5 | 8.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 18.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 33.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.2 | 4.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 2.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 3.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 8.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 35.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 6.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 59.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 3.0 | 41.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.6 | 24.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.4 | 35.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.8 | 29.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 8.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 7.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.4 | 4.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 27.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 18.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 8.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 8.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 16.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 7.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 21.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.2 | 2.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 4.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 2.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 15.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.2 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |