Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
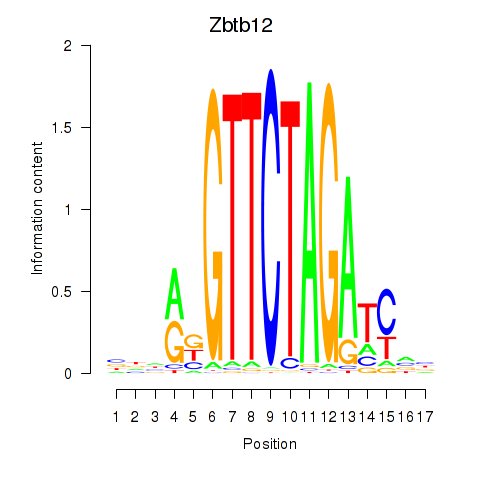
Results for Zbtb12
Z-value: 0.67
Transcription factors associated with Zbtb12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb12
|
ENSRNOG00000000418 | zinc finger and BTB domain containing 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb12 | rn6_v1_chr20_+_4572100_4572100 | -0.23 | 4.9e-05 | Click! |
Activity profile of Zbtb12 motif
Sorted Z-values of Zbtb12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_39164916 | 31.74 |
ENSRNOT00000011309
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr20_+_19325121 | 29.20 |
ENSRNOT00000058151
|
Phyhipl
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr1_+_213597478 | 28.94 |
ENSRNOT00000017562
|
Odf3
|
outer dense fiber of sperm tails 3 |
| chr5_-_17399801 | 24.96 |
ENSRNOT00000011900
|
RGD1563405
|
similar to protein tyrosine phosphatase 4a2 |
| chr20_+_5619012 | 23.36 |
ENSRNOT00000000577
|
Ggnbp1
|
gametogenetin binding protein 1 |
| chr18_+_68983545 | 23.23 |
ENSRNOT00000085317
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chr10_+_88356615 | 21.95 |
ENSRNOT00000022687
|
Klhl10
|
kelch-like family member 10 |
| chr16_-_7007051 | 19.25 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr5_-_59165160 | 18.69 |
ENSRNOT00000029035
|
Fam221b
|
family with sequence similarity 221, member B |
| chr12_+_17614632 | 18.57 |
ENSRNOT00000031896
|
Prkar1b
|
protein kinase cAMP-dependent type 1 regulatory subunit beta |
| chr1_-_227486976 | 18.09 |
ENSRNOT00000035812
|
Ms4a14
|
membrane spanning 4-domains A14 |
| chr8_+_119121669 | 17.27 |
ENSRNOT00000028479
|
Prss46
|
protease, serine, 46 |
| chr15_-_37409668 | 17.20 |
ENSRNOT00000011742
|
Gjb6
|
gap junction protein, beta 6 |
| chr17_-_56269242 | 17.00 |
ENSRNOT00000021901
|
Lyzl1
|
lysozyme-like 1 |
| chr1_+_221443896 | 16.73 |
ENSRNOT00000073942
|
Tmem262
|
transmembrane protein 262 |
| chr2_-_265300868 | 15.84 |
ENSRNOT00000066024
ENSRNOT00000016073 ENSRNOT00000033502 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr16_+_9907598 | 15.82 |
ENSRNOT00000027352
|
Ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr19_+_26416818 | 15.38 |
ENSRNOT00000040473
|
AABR07043200.1
|
|
| chr7_+_129595192 | 15.37 |
ENSRNOT00000071151
|
Zdhhc25
|
zinc finger, DHHC-type containing 25 |
| chr1_-_225035687 | 15.26 |
ENSRNOT00000026539
|
Gng3
|
G protein subunit gamma 3 |
| chr19_-_26946044 | 13.56 |
ENSRNOT00000090756
|
AABR07043271.1
|
|
| chr1_-_266333105 | 12.60 |
ENSRNOT00000027093
|
Arl3
|
ADP ribosylation factor like GTPase 3 |
| chr7_+_123482255 | 12.46 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr13_-_79899479 | 12.40 |
ENSRNOT00000035815
|
RGD1309106
|
similar to hypothetical protein |
| chr10_+_90988732 | 11.86 |
ENSRNOT00000003892
|
FAM187A
|
family with sequence similarity 187, member A |
| chr13_+_100214353 | 11.79 |
ENSRNOT00000073693
|
LOC686031
|
hypothetical protein LOC686031 |
| chr20_-_12820466 | 11.16 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr3_+_47677720 | 10.53 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr3_+_101899068 | 10.44 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr18_-_36513317 | 10.10 |
ENSRNOT00000025453
|
Plac8l1
|
PLAC8-like 1 |
| chr15_-_33775109 | 9.72 |
ENSRNOT00000033722
|
Jph4
|
junctophilin 4 |
| chr2_+_187977008 | 9.63 |
ENSRNOT00000081531
ENSRNOT00000092951 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr16_+_51748970 | 9.51 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr1_+_99762253 | 9.42 |
ENSRNOT00000065876
|
Klk6
|
kallikrein related-peptidase 6 |
| chr1_-_141349881 | 9.26 |
ENSRNOT00000021109
|
Rhcg
|
Rh family, C glycoprotein |
| chr7_-_117440342 | 9.13 |
ENSRNOT00000018528
|
Tssk5
|
testis-specific serine kinase 5 |
| chr2_-_236480502 | 9.06 |
ENSRNOT00000015020
|
Sgms2
|
sphingomyelin synthase 2 |
| chr10_-_29026002 | 8.77 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr9_-_81565416 | 8.48 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chrX_-_156540733 | 8.33 |
ENSRNOT00000091029
|
LOC108348261
|
transketolase-like protein 1 |
| chr1_+_166428761 | 7.97 |
ENSRNOT00000085555
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr8_-_96985558 | 7.94 |
ENSRNOT00000018553
|
LOC501033
|
similar to UPF0258 protein KIAA1024 |
| chr18_-_68983619 | 7.67 |
ENSRNOT00000014670
|
LOC361346
|
similar to chromosome 18 open reading frame 54 |
| chr2_+_206314213 | 7.61 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr10_-_88357025 | 7.49 |
ENSRNOT00000080006
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr15_-_39705208 | 7.49 |
ENSRNOT00000015412
|
Phf11
|
PHD finger protein 11 |
| chr10_+_89459650 | 7.13 |
ENSRNOT00000028149
ENSRNOT00000077942 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr7_-_92996025 | 6.66 |
ENSRNOT00000076327
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr12_+_37829579 | 6.58 |
ENSRNOT00000084587
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr10_-_88356538 | 5.96 |
ENSRNOT00000022430
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr3_+_55886695 | 5.72 |
ENSRNOT00000009396
|
Bbs5
|
Bardet-Biedl syndrome 5 |
| chr3_+_143109316 | 5.59 |
ENSRNOT00000006555
|
LOC257643
|
cystatin SC |
| chr1_-_146968689 | 5.51 |
ENSRNOT00000023181
|
LOC690082
|
similar to melanoma ubiquitous mutated protein |
| chrX_+_6430594 | 5.50 |
ENSRNOT00000044009
|
Maob
|
monoamine oxidase B |
| chr9_-_14550625 | 5.26 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_+_113984824 | 5.21 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr1_-_216971183 | 5.21 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chrX_+_152196129 | 4.57 |
ENSRNOT00000087679
|
Magea4
|
melanoma antigen, family A, 4 |
| chr11_+_88699222 | 4.46 |
ENSRNOT00000084177
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_-_91518996 | 4.38 |
ENSRNOT00000005835
|
Pole2
|
DNA polymerase epsilon 2, accessory subunit |
| chr2_-_250862419 | 4.30 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr10_-_45579029 | 4.28 |
ENSRNOT00000080028
|
Arf1
|
ADP-ribosylation factor 1 |
| chr13_+_90692666 | 4.23 |
ENSRNOT00000010028
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr6_-_125853461 | 4.12 |
ENSRNOT00000007505
|
Atxn3
|
ataxin 3 |
| chr19_-_37819789 | 3.71 |
ENSRNOT00000036702
|
Cenpt
|
centromere protein T |
| chrX_-_112328642 | 3.52 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr14_+_32257805 | 3.40 |
ENSRNOT00000039626
|
AABR07014804.1
|
|
| chrX_+_156909913 | 3.31 |
ENSRNOT00000084390
|
L1cam
|
L1 cell adhesion molecule |
| chr2_-_23909016 | 3.18 |
ENSRNOT00000084044
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr10_-_49009156 | 2.98 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr15_-_28935269 | 2.96 |
ENSRNOT00000060427
|
Olr1645
|
olfactory receptor 1645 |
| chr13_+_71656651 | 2.79 |
ENSRNOT00000050365
|
Zfp648
|
zinc finger protein 648 |
| chr7_+_101069104 | 2.77 |
ENSRNOT00000056894
|
AABR07058124.2
|
|
| chr1_-_226292480 | 2.54 |
ENSRNOT00000039133
|
Myrf
|
myelin regulatory factor |
| chr1_+_198409360 | 2.53 |
ENSRNOT00000013691
ENSRNOT00000091295 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr5_-_166926636 | 2.24 |
ENSRNOT00000023287
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr15_-_61772516 | 2.21 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr6_+_22296128 | 2.12 |
ENSRNOT00000087805
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_+_59765835 | 2.02 |
ENSRNOT00000014842
|
Fpr1
|
formyl peptide receptor 1 |
| chr8_-_73122740 | 1.93 |
ENSRNOT00000012026
|
Tln2
|
talin 2 |
| chr12_-_24365324 | 1.90 |
ENSRNOT00000032250
|
Trim50
|
tripartite motif-containing 50 |
| chr3_+_100787449 | 1.82 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr12_+_47074200 | 1.82 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr1_-_144601327 | 1.80 |
ENSRNOT00000029244
ENSRNOT00000078144 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr6_+_9790422 | 1.77 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chr14_-_108412823 | 1.74 |
ENSRNOT00000081405
|
Pex13
|
peroxisomal biogenesis factor 13 |
| chr6_+_129609074 | 1.72 |
ENSRNOT00000064957
|
Papola
|
poly (A) polymerase alpha |
| chr1_+_59704998 | 1.60 |
ENSRNOT00000014598
|
LOC688452
|
hypothetical protein LOC688452 |
| chr3_-_23353425 | 1.49 |
ENSRNOT00000019703
|
Golga1
|
golgin A1 |
| chr1_-_199320727 | 1.47 |
ENSRNOT00000022452
|
Zfp668
|
zinc finger protein 668 |
| chr2_+_123734199 | 1.40 |
ENSRNOT00000038729
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr7_-_12598370 | 1.40 |
ENSRNOT00000026708
|
Arid3a
|
AT-rich interaction domain 3A |
| chr6_+_145740035 | 1.39 |
ENSRNOT00000064868
|
Cdca7l
|
cell division cycle associated 7 like |
| chr1_+_85143789 | 1.39 |
ENSRNOT00000071691
|
Acp7
|
acid phosphatase 7, tartrate resistant |
| chr6_-_36940868 | 1.36 |
ENSRNOT00000006470
|
Gen1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr3_+_100786862 | 1.30 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr10_-_55589978 | 0.87 |
ENSRNOT00000007087
|
LOC100912917
|
phosphoribosylformylglycinamidine synthase-like |
| chr12_-_19254527 | 0.87 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr6_+_24377327 | 0.71 |
ENSRNOT00000043641
|
Lclat1
|
lysocardiolipin acyltransferase 1 |
| chr7_-_120780641 | 0.69 |
ENSRNOT00000076164
|
Ddx17
|
DEAD-box helicase 17 |
| chr7_-_140580783 | 0.54 |
ENSRNOT00000087327
|
Dhh
|
desert hedgehog |
| chr5_+_139385429 | 0.53 |
ENSRNOT00000078622
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr1_-_78626284 | 0.45 |
ENSRNOT00000052299
|
LOC108349752
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chr11_-_29710849 | 0.41 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr1_+_189328246 | 0.38 |
ENSRNOT00000084260
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr1_-_224698514 | 0.29 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr10_+_86399827 | 0.25 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr5_-_58106706 | 0.21 |
ENSRNOT00000019561
|
Dctn3
|
dynactin subunit 3 |
| chr8_-_36720810 | 0.06 |
ENSRNOT00000089745
|
Gm1113
|
predicted gene 1113 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.2 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 3.7 | 22.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 3.1 | 9.3 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 2.4 | 9.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 1.5 | 18.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.4 | 4.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 1.3 | 10.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.3 | 9.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.2 | 3.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.0 | 8.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.9 | 4.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.8 | 3.3 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.7 | 5.5 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.6 | 4.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.6 | 9.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.6 | 19.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.5 | 23.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.4 | 1.8 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.4 | 2.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 3.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.4 | 5.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 14.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.3 | 4.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 9.4 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 | 8.8 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 3.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 1.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 0.5 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 2.5 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 2.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 2.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 1.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 1.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 7.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 17.2 | GO:0042471 | ear morphogenesis(GO:0042471) |
| 0.1 | 2.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 10.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 8.5 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 5.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 15.8 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 14.9 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 7.6 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 2.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 5.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 5.3 | GO:0042278 | purine nucleoside metabolic process(GO:0042278) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 11.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 2.1 | 18.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.5 | 28.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.1 | 9.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.0 | 3.1 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.9 | 4.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.8 | 17.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 8.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 15.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.6 | 5.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.4 | 15.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 3.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.3 | 4.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.3 | 12.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.3 | 23.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.3 | 8.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 3.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 7.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 31.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 9.6 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 4.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 3.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 12.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 2.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 3.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 5.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 4.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 7.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 18.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 2.2 | 11.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.8 | 9.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.5 | 15.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 1.3 | 17.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.8 | 13.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 4.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 23.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.5 | 5.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 1.8 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.4 | 9.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 3.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.4 | 4.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.3 | 2.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 10.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 2.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.3 | 8.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 3.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 4.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 1.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 14.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 12.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 19.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 4.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 10.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 5.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 23.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 10.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 4.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 8.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 5.2 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 8.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 15.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 9.1 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 22.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.4 | 4.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 14.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 9.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 7.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 19.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 17.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.8 | 18.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.7 | 23.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.7 | 10.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 15.3 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.4 | 4.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 4.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 9.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 14.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 8.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 10.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 5.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.8 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.1 | 3.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 8.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 3.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 2.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |