Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Xbp1_Creb3l1
Z-value: 1.16
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
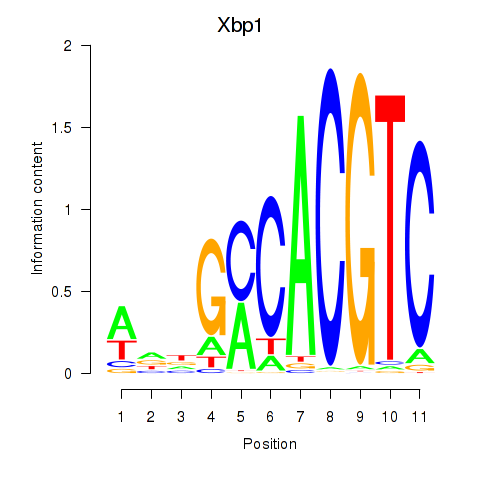
Xbp1
|
ENSRNOG00000010298 | X-box binding protein 1 |
|
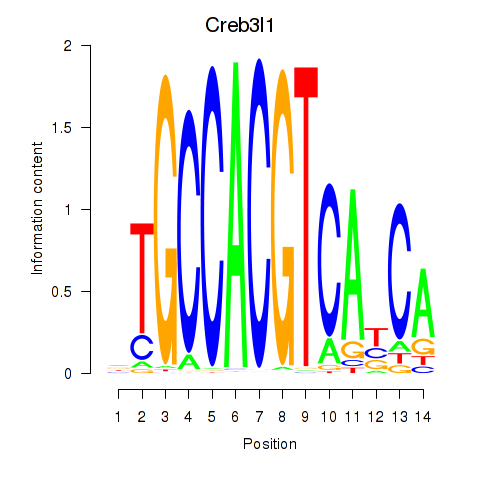
Creb3l1
|
ENSRNOG00000005413 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | rn6_v1_chr3_-_80933283_80933283 | 0.56 | 5.8e-28 | Click! |
| Xbp1 | rn6_v1_chr14_+_85753760_85753760 | 0.42 | 2.6e-15 | Click! |
Activity profile of Xbp1_Creb3l1 motif
Sorted Z-values of Xbp1_Creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_101086198 | 52.35 |
ENSRNOT00000027917
|
Rcn3
|
reticulocalbin 3 |
| chr1_-_101085884 | 49.81 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr2_+_227455722 | 46.48 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr18_+_52423883 | 32.44 |
ENSRNOT00000022017
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr7_+_120749752 | 31.00 |
ENSRNOT00000087888
|
Kdelr3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr4_+_56625561 | 29.46 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr5_-_173081839 | 29.32 |
ENSRNOT00000066880
|
Mmp23
|
matrix metallopeptidase 23 |
| chr18_-_47513030 | 28.00 |
ENSRNOT00000083881
ENSRNOT00000074226 |
Lox
|
lysyl oxidase |
| chr17_+_26925828 | 24.66 |
ENSRNOT00000018310
|
Txndc5
|
thioredoxin domain containing 5 |
| chr13_+_80125391 | 21.64 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr3_-_138116664 | 18.04 |
ENSRNOT00000055602
|
Rrbp1
|
ribosome binding protein 1 |
| chr17_+_9736786 | 17.78 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr7_+_11490852 | 16.88 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr7_+_31784438 | 15.97 |
ENSRNOT00000010914
ENSRNOT00000010929 |
Ikbip
|
IKBKB interacting protein |
| chr4_+_119997268 | 15.81 |
ENSRNOT00000073429
|
Rpn1
|
ribophorin I |
| chr11_+_46009322 | 15.46 |
ENSRNOT00000002233
|
Tmem45a
|
transmembrane protein 45A |
| chr5_-_59025631 | 15.45 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr11_+_46714355 | 15.41 |
ENSRNOT00000073404
|
Tmem45al
|
transmembrane protein 45A-like |
| chr11_+_68493035 | 14.26 |
ENSRNOT00000067984
|
Pdia5
|
protein disulfide isomerase family A, member 5 |
| chr9_-_100624638 | 14.13 |
ENSRNOT00000051155
|
Hdlbp
|
high density lipoprotein binding protein |
| chr6_+_53401109 | 13.99 |
ENSRNOT00000014763
|
Twist1
|
twist family bHLH transcription factor 1 |
| chr5_+_167952728 | 13.39 |
ENSRNOT00000085315
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr1_+_251145253 | 12.78 |
ENSRNOT00000083555
ENSRNOT00000014979 |
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr3_+_95232166 | 12.24 |
ENSRNOT00000017952
|
LOC691083
|
hypothetical protein LOC691083 |
| chr8_+_112594691 | 12.06 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr1_-_226152524 | 11.93 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr15_+_57891680 | 11.55 |
ENSRNOT00000001383
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr5_-_59063592 | 11.47 |
ENSRNOT00000022400
|
Tln1
|
talin 1 |
| chr4_-_120467932 | 11.36 |
ENSRNOT00000018454
|
Sec61a1
|
Sec61 translocon alpha 1 subunit |
| chr5_+_76150840 | 11.33 |
ENSRNOT00000020702
|
Dnajc25
|
DnaJ heat shock protein family (Hsp40) member C25 |
| chr17_+_30965942 | 11.20 |
ENSRNOT00000060143
|
Pxdc1
|
PX domain containing 1 |
| chr1_-_185143272 | 11.14 |
ENSRNOT00000027752
|
Nucb2
|
nucleobindin 2 |
| chr10_+_39435227 | 11.05 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr20_+_46250363 | 10.75 |
ENSRNOT00000076522
ENSRNOT00000000334 |
Cd164
|
CD164 molecule |
| chr5_+_5215196 | 10.50 |
ENSRNOT00000010481
|
Tram1
|
translocation associated membrane protein 1 |
| chr1_+_220470089 | 9.80 |
ENSRNOT00000027406
ENSRNOT00000078657 |
Yif1a
|
Yip1 interacting factor homolog A, membrane trafficking protein |
| chr4_-_82258765 | 9.69 |
ENSRNOT00000008523
|
Hoxa5
|
homeo box A5 |
| chr16_+_2338333 | 9.60 |
ENSRNOT00000017692
|
Arf4
|
ADP-ribosylation factor 4 |
| chr1_+_101859346 | 9.26 |
ENSRNOT00000028627
|
Kdelr1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr9_-_82699551 | 9.20 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr20_-_3822754 | 8.95 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr8_-_97083120 | 8.78 |
ENSRNOT00000018603
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr12_-_13210758 | 8.64 |
ENSRNOT00000001438
|
Kdelr2
|
KDEL endoplasmic reticulum protein retention receptor 2 |
| chr5_+_63192298 | 8.48 |
ENSRNOT00000008329
|
Sec61b
|
Sec61 translocon beta subunit |
| chr5_-_58445953 | 8.31 |
ENSRNOT00000046102
|
Vcp
|
valosin-containing protein |
| chr4_-_84768249 | 8.21 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
| chr1_+_199941161 | 7.85 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr4_+_55772377 | 7.75 |
ENSRNOT00000047698
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chrX_-_156999650 | 7.55 |
ENSRNOT00000083557
|
Ssr4
|
signal sequence receptor subunit 4 |
| chr8_-_13109487 | 7.43 |
ENSRNOT00000061616
|
Amotl1
|
angiomotin-like 1 |
| chr7_+_24939498 | 7.18 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr20_+_47494270 | 7.17 |
ENSRNOT00000036365
|
Sec63
|
SEC63 homolog, protein translocation regulator |
| chr20_+_47494424 | 7.10 |
ENSRNOT00000087718
|
Sec63
|
SEC63 homolog, protein translocation regulator |
| chr10_-_37209881 | 7.08 |
ENSRNOT00000090475
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr9_-_100592271 | 7.05 |
ENSRNOT00000086433
|
Hdlbp
|
high density lipoprotein binding protein |
| chr1_+_225096598 | 6.74 |
ENSRNOT00000035650
|
Ganab
|
glucosidase II alpha subunit |
| chr11_-_64752544 | 6.70 |
ENSRNOT00000048738
|
Tmem39a
|
transmembrane protein 39a |
| chr16_-_71125316 | 6.68 |
ENSRNOT00000020900
|
Plpp5
|
phospholipid phosphatase 5 |
| chr10_+_65448950 | 6.64 |
ENSRNOT00000082348
ENSRNOT00000037016 |
Rab34
|
RAB34, member RAS oncogene family |
| chr10_-_105149971 | 6.56 |
ENSRNOT00000012436
|
Srp68
|
signal recognition particle 68 |
| chr11_+_64601029 | 6.40 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr17_-_1830001 | 6.31 |
ENSRNOT00000066230
|
Aaed1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr7_-_2819299 | 6.11 |
ENSRNOT00000045547
|
Slc39a5
|
solute carrier family 39 member 5 |
| chr8_-_39243882 | 5.91 |
ENSRNOT00000082086
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr11_-_66819079 | 5.81 |
ENSRNOT00000003255
|
Golgb1
|
golgin B1 |
| chr12_-_37480297 | 5.77 |
ENSRNOT00000001397
|
Tmed2
|
transmembrane p24 trafficking protein 2 |
| chr11_-_51202703 | 5.74 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr8_-_39243705 | 5.71 |
ENSRNOT00000043518
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr2_+_9526209 | 5.62 |
ENSRNOT00000021818
|
Lysmd3
|
LysM domain containing 3 |
| chr10_-_105228547 | 5.43 |
ENSRNOT00000028875
|
Srp68
|
signal recognition particle 68 |
| chr19_-_38120578 | 5.36 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr17_+_82065937 | 5.01 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr19_-_37938857 | 4.96 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr6_+_91476698 | 4.88 |
ENSRNOT00000005608
|
Mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr1_+_164425475 | 4.86 |
ENSRNOT00000023386
|
Klhl35
|
kelch-like family member 35 |
| chr4_-_29092753 | 4.72 |
ENSRNOT00000014810
|
Bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr16_-_20873344 | 4.68 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr1_+_221644867 | 4.61 |
ENSRNOT00000054830
|
Ehd1
|
EH-domain containing 1 |
| chr16_+_19501193 | 4.54 |
ENSRNOT00000085328
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr20_+_22728208 | 4.43 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr2_+_85377318 | 4.29 |
ENSRNOT00000016506
ENSRNOT00000085094 |
Sema5a
|
semaphorin 5A |
| chr20_+_31298269 | 4.28 |
ENSRNOT00000000673
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr7_-_97759852 | 4.24 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr2_+_198823366 | 4.23 |
ENSRNOT00000083769
ENSRNOT00000028814 |
Pias3
|
protein inhibitor of activated STAT, 3 |
| chr14_-_17436897 | 4.21 |
ENSRNOT00000003277
|
Uso1
|
USO1 vesicle transport factor |
| chr5_-_24631679 | 4.11 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chrX_-_34162827 | 3.95 |
ENSRNOT00000046759
|
RGD1560203
|
similar to ferritin heavy polypeptide-like 17 |
| chr8_-_49075892 | 3.82 |
ENSRNOT00000082687
|
Arcn1
|
archain 1 |
| chr4_+_113948514 | 3.79 |
ENSRNOT00000011521
|
LOC103692171
|
mannosyl-oligosaccharide glucosidase |
| chr4_-_145699577 | 3.73 |
ENSRNOT00000014377
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr15_-_34322115 | 3.71 |
ENSRNOT00000026734
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr13_+_89253617 | 3.61 |
ENSRNOT00000044575
|
LOC100362384
|
ferritin light chain 1-like |
| chr19_+_39357791 | 3.35 |
ENSRNOT00000086435
ENSRNOT00000015006 |
Cyb5b
|
cytochrome b5 type B |
| chr8_+_79606789 | 3.35 |
ENSRNOT00000087114
|
Pygo1
|
pygopus family PHD finger 1 |
| chr17_+_82066152 | 3.06 |
ENSRNOT00000083034
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chr1_-_190965115 | 2.99 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr15_-_45927804 | 2.98 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr5_+_59063531 | 2.96 |
ENSRNOT00000085311
ENSRNOT00000065909 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr7_+_29282305 | 2.93 |
ENSRNOT00000007623
|
Arl1
|
ADP-ribosylation factor like GTPase 1 |
| chr1_-_260517323 | 2.80 |
ENSRNOT00000018043
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr12_+_47218969 | 2.80 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr2_-_183582553 | 2.72 |
ENSRNOT00000014236
|
Arfip1
|
ADP-ribosylation factor interacting protein 1 |
| chr3_+_56966654 | 2.71 |
ENSRNOT00000088097
|
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr4_+_118852062 | 2.70 |
ENSRNOT00000090827
ENSRNOT00000025070 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr7_-_124198200 | 2.66 |
ENSRNOT00000078947
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr8_+_62341613 | 2.65 |
ENSRNOT00000066923
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr5_-_117612123 | 2.53 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr3_-_95418679 | 2.51 |
ENSRNOT00000018074
|
Rcn1
|
reticulocalbin 1 |
| chrX_+_73390903 | 2.44 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr9_+_100080144 | 2.38 |
ENSRNOT00000071411
|
Rnpepl1
|
arginyl aminopeptidase like 1 |
| chr1_+_266530477 | 2.36 |
ENSRNOT00000054699
|
Cnnm2
|
cyclin and CBS domain divalent metal cation transport mediator 2 |
| chr9_-_82673898 | 2.34 |
ENSRNOT00000027165
|
Chpf
|
chondroitin polymerizing factor |
| chr5_-_152920343 | 2.31 |
ENSRNOT00000022956
|
Man1c1
|
mannosidase, alpha, class 1C, member 1 |
| chr1_-_6970040 | 2.24 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr18_+_1504178 | 2.11 |
ENSRNOT00000048198
|
Rpl36al
|
ribosomal protein L36a-like |
| chr13_-_90467235 | 2.09 |
ENSRNOT00000081534
ENSRNOT00000008150 |
Ncstn
|
nicastrin |
| chr7_-_31784192 | 2.01 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr12_-_12407561 | 1.98 |
ENSRNOT00000037251
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr7_-_24668250 | 1.94 |
ENSRNOT00000083806
|
Tmem263
|
transmembrane protein 263 |
| chrX_+_9815652 | 1.93 |
ENSRNOT00000091382
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr15_-_57805184 | 1.92 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr1_-_141188031 | 1.83 |
ENSRNOT00000044567
|
Polg
|
DNA polymerase gamma, catalytic subunit |
| chr19_-_37210412 | 1.72 |
ENSRNOT00000083097
|
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr9_-_61810417 | 1.66 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr2_-_2891253 | 1.64 |
ENSRNOT00000029250
|
Arsk
|
arylsulfatase family, member K |
| chr16_-_49484985 | 1.55 |
ENSRNOT00000016723
|
Ufsp2
|
UFM1-specific peptidase 2 |
| chr3_+_11317183 | 1.51 |
ENSRNOT00000091171
ENSRNOT00000016341 |
Golga2
|
golgin A2 |
| chr7_-_11406771 | 1.50 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr3_-_10269693 | 1.50 |
ENSRNOT00000021871
|
Fubp3
|
far upstream element binding protein 3 |
| chr15_-_4057104 | 1.49 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr10_-_84789832 | 1.47 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr5_-_39650894 | 1.47 |
ENSRNOT00000010421
ENSRNOT00000089522 |
Ufl1
|
Ufm1-specific ligase 1 |
| chr7_+_66717267 | 1.39 |
ENSRNOT00000005654
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr1_-_220067123 | 1.21 |
ENSRNOT00000071020
ENSRNOT00000072373 |
Rbm14
|
RNA binding motif protein 14 |
| chr6_+_21051327 | 1.21 |
ENSRNOT00000052131
|
Fam98a
|
family with sequence similarity 98, member A |
| chr16_+_20873642 | 1.12 |
ENSRNOT00000004891
|
Ddx49
|
DEAD-box helicase 49 |
| chr1_-_87174530 | 1.10 |
ENSRNOT00000089578
|
Kcnk6
|
potassium two pore domain channel subfamily K member 6 |
| chr3_+_14889510 | 0.98 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr6_+_38474804 | 0.95 |
ENSRNOT00000009779
|
Nbas
|
neuroblastoma amplified sequence |
| chr4_+_342302 | 0.92 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr2_-_30846149 | 0.91 |
ENSRNOT00000025506
|
Slc30a5
|
solute carrier family 30 member 5 |
| chr7_-_11444786 | 0.82 |
ENSRNOT00000027323
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr17_-_85141210 | 0.79 |
ENSRNOT00000000162
|
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr2_-_211352540 | 0.77 |
ENSRNOT00000023728
|
LOC108349010
|
protein kish-B |
| chr14_+_38192870 | 0.73 |
ENSRNOT00000077080
|
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chrX_+_45637415 | 0.72 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr9_+_61810859 | 0.66 |
ENSRNOT00000079058
|
AABR07067763.1
|
|
| chr5_-_152227677 | 0.65 |
ENSRNOT00000020051
|
Dhdds
|
dehydrodolichyl diphosphate synthase subunit |
| chr3_+_160945359 | 0.64 |
ENSRNOT00000019761
|
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr13_+_90467265 | 0.45 |
ENSRNOT00000008518
|
Copa
|
coatomer protein complex subunit alpha |
| chr7_+_117951154 | 0.34 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr12_-_52558606 | 0.28 |
ENSRNOT00000056656
|
Golga3
|
golgin A3 |
| chr10_-_16689321 | 0.21 |
ENSRNOT00000028173
|
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr8_+_5790034 | 0.20 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr10_-_56558487 | 0.20 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr11_-_16889201 | 0.16 |
ENSRNOT00000002121
|
Btg3
|
BTG anti-proliferation factor 3 |
| chr10_-_59360661 | 0.14 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr1_+_241501679 | 0.08 |
ENSRNOT00000054775
|
Ptar1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr5_+_62109328 | 0.04 |
ENSRNOT00000012260
|
Nans
|
N-acetylneuraminate synthase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Xbp1_Creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 7.0 | 48.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 5.9 | 17.8 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 4.7 | 14.0 | GO:0035359 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 3.9 | 11.6 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 3.7 | 11.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 3.3 | 13.4 | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 3.2 | 9.7 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 2.8 | 28.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 2.8 | 8.3 | GO:2000158 | flavin-containing compound metabolic process(GO:0042726) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 2.5 | 7.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 2.3 | 11.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 2.0 | 6.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.8 | 29.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 1.8 | 11.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.8 | 12.8 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.8 | 12.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 1.7 | 11.6 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 1.5 | 4.6 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 1.5 | 4.5 | GO:0036101 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 1.3 | 3.8 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 1.1 | 16.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 1.1 | 12.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.0 | 11.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.0 | 3.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 1.0 | 5.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.9 | 9.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 85.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.7 | 6.6 | GO:0044351 | macropinocytosis(GO:0044351) phagosome-lysosome fusion(GO:0090385) |
| 0.7 | 11.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.7 | 2.7 | GO:1901073 | glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.6 | 5.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.6 | 37.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.5 | 4.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.5 | 6.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.5 | 14.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.4 | 2.5 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.4 | 5.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.4 | 7.9 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 4.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.4 | 9.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.4 | 2.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.4 | 4.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.3 | 9.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.3 | 1.0 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.3 | 1.8 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.3 | 1.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.3 | 2.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 15.4 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.2 | 0.7 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.2 | 2.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 2.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 2.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 21.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.2 | 10.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 2.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 | 6.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 7.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 3.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.2 | 0.9 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 2.0 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 6.1 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.1 | 2.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 3.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 25.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 6.3 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 0.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 2.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 5.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 9.0 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 22.0 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.2 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 29.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 3.9 | 55.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 3.2 | 16.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 2.9 | 11.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.8 | 8.3 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 2.8 | 11.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 2.2 | 15.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.6 | 27.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.5 | 4.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.5 | 9.2 | GO:1990393 | 3M complex(GO:1990393) |
| 1.5 | 30.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 1.4 | 13.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.9 | 10.1 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.8 | 7.7 | GO:0097433 | dense body(GO:0097433) |
| 0.7 | 21.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.7 | 19.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.6 | 1.8 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.5 | 37.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.5 | 21.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 2.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.4 | 2.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.4 | 28.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 1.0 | GO:1990032 | climbing fiber(GO:0044301) parallel fiber(GO:1990032) |
| 0.3 | 0.9 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.3 | 0.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 10.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 6.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 148.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 3.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 3.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 6.7 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.1 | 2.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 7.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 13.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 21.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 7.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 12.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 15.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 4.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 7.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 6.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 8.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 7.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 14.4 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.8 | 48.9 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 5.6 | 28.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 4.3 | 12.8 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 4.0 | 12.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 3.0 | 12.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 3.0 | 11.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 2.8 | 8.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 2.8 | 11.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 2.1 | 8.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 2.0 | 27.4 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.9 | 38.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.5 | 4.5 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 1.4 | 11.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.2 | 19.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 1.1 | 4.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.9 | 2.7 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.9 | 4.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.8 | 7.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.7 | 9.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 8.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 15.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 5.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 15.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 14.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.4 | 6.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 6.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.3 | 6.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 1.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 4.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.3 | 9.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 11.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 29.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 3.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.2 | 22.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 157.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 2.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 2.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 2.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 19.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 6.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 2.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 2.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 4.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 4.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 6.7 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 6.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 4.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 1.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 11.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 3.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 9.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 9.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 33.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 9.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 2.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 14.9 | GO:0042802 | identical protein binding(GO:0042802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.4 | 4.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.4 | 11.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 14.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 57.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 13.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 3.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 2.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 3.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 4.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 51.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.5 | 70.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 1.2 | 11.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.1 | 6.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 1.0 | 24.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.0 | 8.9 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.0 | 12.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.8 | 17.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 16.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.8 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.5 | 59.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.4 | 15.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 7.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 5.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 18.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.2 | 4.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.2 | 16.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 5.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 1.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.0 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 5.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 6.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 4.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |