Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
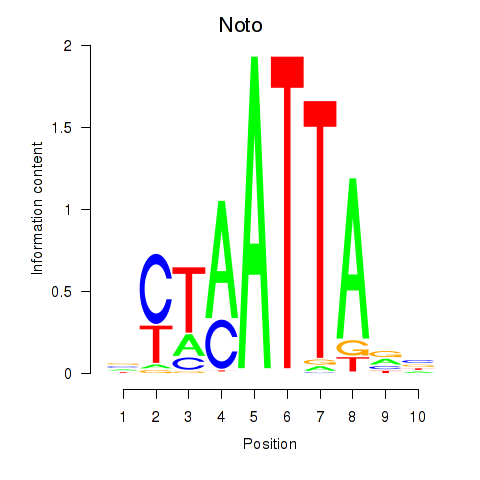
Results for Vsx1_Uncx_Prrx2_Shox2_Noto
Z-value: 0.74
Transcription factors associated with Vsx1_Uncx_Prrx2_Shox2_Noto
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
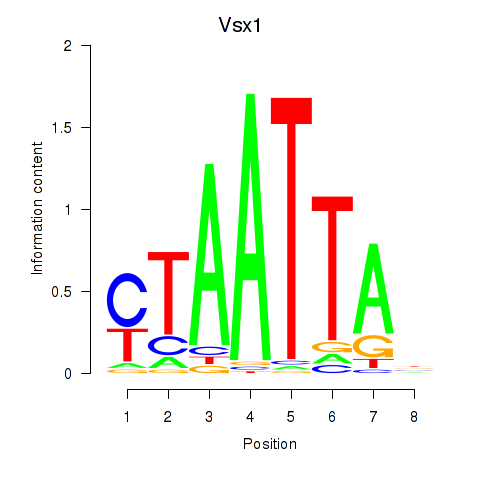
Vsx1
|
ENSRNOG00000042220 | visual system homeobox 1 |
|
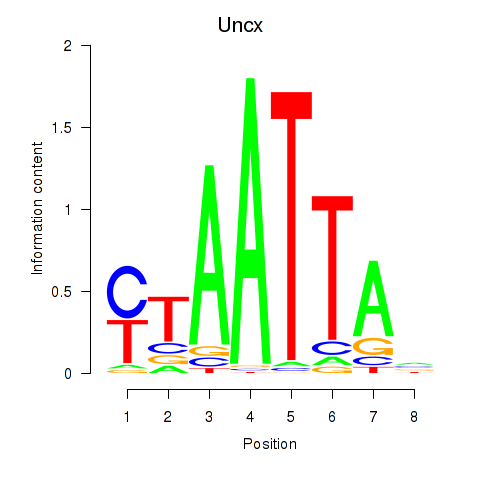
Uncx
|
ENSRNOG00000001284 | UNC homeobox |
|
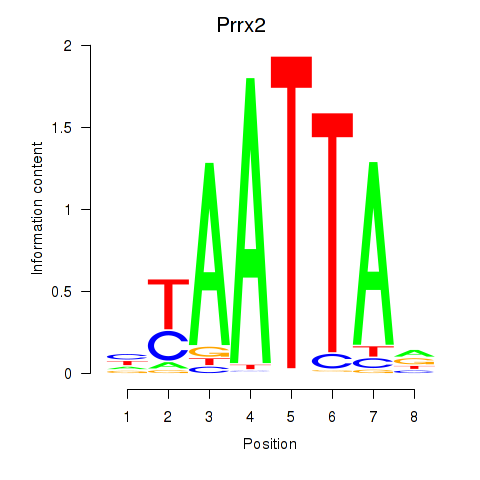
Prrx2
|
ENSRNOG00000058329 | paired related homeobox 2 |
|
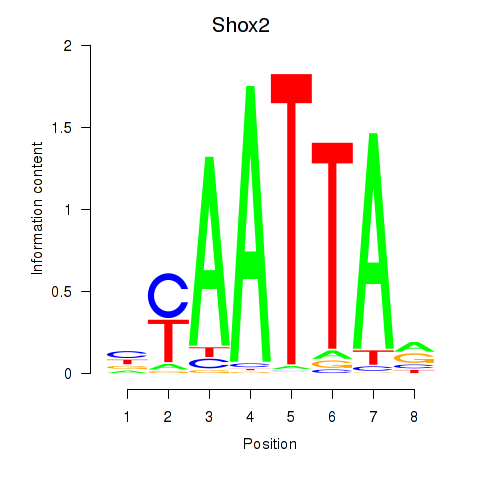
Shox2
|
ENSRNOG00000012478 | short stature homeobox 2 |
|
Noto
|
ENSRNOG00000037863 | notochord homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vsx1 | rn6_v1_chr3_-_146491837_146491837 | -0.38 | 1.8e-12 | Click! |
| Prrx2 | rn6_v1_chr3_+_9681871_9681871 | -0.16 | 4.5e-03 | Click! |
| Noto | rn6_v1_chr4_+_117206355_117206355 | 0.10 | 6.7e-02 | Click! |
| Shox2 | rn6_v1_chr2_-_164126783_164126783 | 0.07 | 2.2e-01 | Click! |
| Uncx | rn6_v1_chr12_-_17186679_17186679 | 0.01 | 9.1e-01 | Click! |
Activity profile of Vsx1_Uncx_Prrx2_Shox2_Noto motif
Sorted Z-values of Vsx1_Uncx_Prrx2_Shox2_Noto motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_53740841 | 55.47 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr7_-_49741540 | 37.08 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr10_-_87232723 | 35.12 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr1_+_105284753 | 32.41 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr16_+_10267482 | 29.21 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr15_+_1054937 | 28.87 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr7_-_69982592 | 28.74 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr1_+_279633671 | 23.23 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr1_+_107344904 | 21.14 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr11_+_58624198 | 18.57 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr4_-_55011415 | 18.32 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr9_-_30844199 | 18.21 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr13_-_76049363 | 15.88 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr18_+_59748444 | 15.64 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr10_-_88670430 | 15.34 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr1_-_173764246 | 15.27 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr7_+_133856101 | 15.12 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr10_-_87286387 | 14.62 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr10_-_87248572 | 14.49 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr7_-_49732764 | 13.43 |
ENSRNOT00000006453
|
Myf5
|
myogenic factor 5 |
| chr7_+_78558701 | 13.23 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_143353925 | 13.01 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr3_+_54253949 | 12.83 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr7_-_143408276 | 12.28 |
ENSRNOT00000013122
ENSRNOT00000091540 |
Krt73
|
keratin 73 |
| chr8_-_39551700 | 11.96 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_+_185863043 | 11.71 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr2_+_145174876 | 11.53 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr9_+_53013413 | 11.33 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr1_+_198383201 | 11.26 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr9_-_85243001 | 11.13 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr1_+_59156251 | 11.13 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr13_+_49005405 | 10.82 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr8_+_5790034 | 10.59 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr3_+_95715193 | 10.04 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr4_-_172063391 | 9.80 |
ENSRNOT00000010158
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr3_-_165537940 | 9.57 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr7_-_143967484 | 9.31 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr12_-_2174131 | 9.25 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_+_47677720 | 9.08 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr4_-_80395502 | 8.61 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr8_+_33239139 | 8.54 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_-_143966863 | 8.35 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr5_+_139790395 | 8.30 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr10_+_54352270 | 8.22 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr11_+_28780446 | 8.22 |
ENSRNOT00000072546
|
Krtap15-1
|
keratin associated protein 15-1 |
| chr10_-_74679858 | 8.01 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr17_+_11683862 | 7.74 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr16_+_46731403 | 7.65 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_276228574 | 7.64 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr10_-_87521514 | 7.63 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chrX_+_118197217 | 7.20 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr2_-_192027225 | 7.09 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr18_-_13183263 | 6.74 |
ENSRNOT00000050933
|
Ccdc178
|
coiled-coil domain containing 178 |
| chr16_+_22361998 | 6.71 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr14_+_96499520 | 6.64 |
ENSRNOT00000074692
|
AABR07016310.1
|
|
| chr2_+_196334626 | 6.49 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr10_-_87468711 | 6.37 |
ENSRNOT00000039983
|
Krtap3-3
|
keratin associated protein 3-3 |
| chr7_+_143122269 | 6.22 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr19_+_39229754 | 6.19 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr7_+_44009069 | 6.15 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr14_-_2032593 | 6.08 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr3_-_37803112 | 5.85 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr14_+_42714315 | 5.74 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr8_+_49441106 | 5.63 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr16_-_24951612 | 5.60 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr9_-_53315915 | 5.50 |
ENSRNOT00000038093
|
Mstn
|
myostatin |
| chr2_+_136993208 | 5.43 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr16_+_29674793 | 5.30 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr2_-_98610368 | 5.13 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr7_-_126382449 | 5.10 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chrX_-_23144324 | 5.09 |
ENSRNOT00000000178
ENSRNOT00000081239 |
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_15278645 | 5.03 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr12_-_47142852 | 5.01 |
ENSRNOT00000001591
|
Pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr6_-_23316962 | 4.82 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr8_+_48718329 | 4.69 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr3_-_52447622 | 4.66 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr9_+_25304876 | 4.65 |
ENSRNOT00000065053
|
Tfap2d
|
transcription factor AP-2 delta |
| chr8_-_83280888 | 4.64 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr6_+_28382962 | 4.60 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr17_-_39824299 | 4.59 |
ENSRNOT00000023412
|
Prl
|
prolactin |
| chr2_-_188559882 | 4.46 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr5_-_28131133 | 4.29 |
ENSRNOT00000067331
|
Slc26a7
|
solute carrier family 26 member 7 |
| chrX_-_110232179 | 4.26 |
ENSRNOT00000014739
|
Serpina7
|
serpin family A member 7 |
| chr1_-_104973648 | 4.23 |
ENSRNOT00000019739
|
Dbx1
|
developing brain homeobox 1 |
| chr10_-_88000423 | 4.21 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr11_+_80742467 | 4.19 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr17_-_79829511 | 4.19 |
ENSRNOT00000085909
|
AABR07028668.1
|
|
| chr6_-_67084234 | 4.16 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr16_+_2634603 | 4.16 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr5_-_28130803 | 4.15 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr6_-_106971250 | 4.13 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr12_-_17186679 | 4.09 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr2_+_18354542 | 4.02 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr8_-_14373894 | 4.00 |
ENSRNOT00000011883
|
Mtnr1b
|
melatonin receptor 1B |
| chr10_+_103395511 | 3.95 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr7_+_136182224 | 3.85 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr13_+_90943255 | 3.78 |
ENSRNOT00000011539
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr3_+_61620192 | 3.76 |
ENSRNOT00000065426
|
Hoxd9
|
homeo box D9 |
| chr11_-_76888178 | 3.68 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr2_+_54466280 | 3.53 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr15_-_27819376 | 3.51 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr14_+_23405717 | 3.50 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr11_-_28777603 | 3.50 |
ENSRNOT00000002132
|
Krtap14
|
keratin associated protein 14 |
| chr3_-_44086006 | 3.48 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr4_-_41212072 | 3.48 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr9_-_14550625 | 3.39 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_+_2752680 | 3.38 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr9_+_73378057 | 3.36 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chrX_+_131381134 | 3.33 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr2_-_139528162 | 3.26 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr4_-_180234804 | 3.09 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr5_-_12526962 | 3.07 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr4_-_4473307 | 3.07 |
ENSRNOT00000045773
|
Dpp6
|
dipeptidyl peptidase like 6 |
| chr10_+_91217079 | 3.05 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr8_-_43304560 | 3.03 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr14_+_23330933 | 3.02 |
ENSRNOT00000088552
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr17_-_45746753 | 2.96 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr1_+_184301493 | 2.96 |
ENSRNOT00000078096
|
Insc
|
inscuteable homolog (Drosophila) |
| chr6_-_36876805 | 2.92 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr10_-_87503591 | 2.87 |
ENSRNOT00000037980
|
Krtap1-1
|
keratin associated protein 1-1 |
| chr19_-_49448072 | 2.87 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr4_-_169036950 | 2.83 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr7_-_145062956 | 2.79 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr2_-_170301348 | 2.77 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr3_+_5709236 | 2.76 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr4_-_58250798 | 2.68 |
ENSRNOT00000048436
|
Klf14
|
Kruppel-like factor 14 |
| chr9_+_73418607 | 2.64 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr16_-_9658484 | 2.61 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr1_+_101599018 | 2.54 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr3_-_11452529 | 2.53 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr4_+_147832136 | 2.51 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr11_-_29710849 | 2.47 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr4_+_57925323 | 2.46 |
ENSRNOT00000085798
|
Cpa5
|
carboxypeptidase A5 |
| chr11_-_43022565 | 2.44 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr1_+_140998240 | 2.38 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr3_-_141411170 | 2.34 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr5_-_136965191 | 2.32 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr2_-_45518502 | 2.28 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr8_-_84506328 | 2.26 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr7_-_106653567 | 2.15 |
ENSRNOT00000042522
|
Oc90
|
otoconin 90 |
| chr8_+_40410604 | 2.14 |
ENSRNOT00000049392
|
Olr1202
|
olfactory receptor 1202 |
| chr4_+_1671269 | 2.14 |
ENSRNOT00000071452
|
Olr1251
|
olfactory receptor 1251 |
| chr4_+_180291389 | 2.13 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr10_-_93675991 | 2.12 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr9_+_95398237 | 2.11 |
ENSRNOT00000025879
|
Trpm8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr9_-_82146874 | 2.11 |
ENSRNOT00000024190
|
Fev
|
FEV, ETS transcription factor |
| chr8_+_122076759 | 2.08 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr2_+_144861455 | 2.02 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr14_+_7630482 | 2.01 |
ENSRNOT00000091581
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr15_+_11298478 | 1.99 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr14_+_4362717 | 1.97 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr1_-_48033343 | 1.92 |
ENSRNOT00000019531
ENSRNOT00000076422 |
Tcp1
|
t-complex 1 |
| chr4_+_2055615 | 1.91 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr18_-_61057365 | 1.89 |
ENSRNOT00000042352
|
Alpk2
|
alpha-kinase 2 |
| chr9_-_18371856 | 1.88 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr6_+_22696397 | 1.87 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chrX_+_6273733 | 1.86 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr1_-_167971151 | 1.86 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr2_+_189993262 | 1.84 |
ENSRNOT00000016034
|
S100a3
|
S100 calcium binding protein A3 |
| chr12_-_29743705 | 1.82 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr13_-_36290531 | 1.82 |
ENSRNOT00000071388
|
Steap3
|
STEAP3 metalloreductase |
| chr10_+_87782376 | 1.78 |
ENSRNOT00000017415
|
LOC680396
|
hypothetical protein LOC680396 |
| chrX_-_16532089 | 1.75 |
ENSRNOT00000093156
|
Dgkk
|
diacylglycerol kinase kappa |
| chrX_-_156589907 | 1.75 |
ENSRNOT00000083147
|
Opn1mw
|
opsin 1 (cone pigments), medium-wave-sensitive |
| chr15_+_12827707 | 1.72 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr3_+_9403840 | 1.72 |
ENSRNOT00000071380
|
NEWGENE_735020
|
pyroglutamylated RFamide peptide |
| chr10_+_16259728 | 1.69 |
ENSRNOT00000028115
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr2_+_18392142 | 1.69 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr5_+_135574172 | 1.68 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr2_-_61692487 | 1.66 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr8_-_53223804 | 1.65 |
ENSRNOT00000008965
|
Htr3a
|
5-hydroxytryptamine receptor 3A |
| chr5_+_36566783 | 1.63 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr11_-_4397361 | 1.63 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr12_-_35979193 | 1.60 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr4_+_108301129 | 1.59 |
ENSRNOT00000007993
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr20_-_30947484 | 1.58 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr6_+_49825469 | 1.53 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr2_-_173087648 | 1.49 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr10_+_64930023 | 1.43 |
ENSRNOT00000071102
|
AABR07030053.1
|
|
| chr3_-_61636038 | 1.42 |
ENSRNOT00000086887
|
AABR07052559.1
|
|
| chr18_+_81694808 | 1.41 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr3_-_10196626 | 1.40 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr13_+_91054974 | 1.40 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr8_+_41350182 | 1.40 |
ENSRNOT00000072823
|
LOC100911758
|
olfactory receptor 143-like |
| chr3_-_77605962 | 1.39 |
ENSRNOT00000090062
|
Olr665
|
olfactory receptor 665 |
| chr1_-_230687157 | 1.39 |
ENSRNOT00000048193
|
Olr380
|
olfactory receptor 380 |
| chr14_-_21252538 | 1.38 |
ENSRNOT00000005003
|
Ambn
|
ameloblastin |
| chr11_-_28900376 | 1.37 |
ENSRNOT00000061606
|
Krtap16-5
|
keratin associated protein 16-5 |
| chr20_-_3728844 | 1.36 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr9_-_79630452 | 1.35 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr15_+_4064706 | 1.35 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr4_+_68656928 | 1.35 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr1_+_80141630 | 1.32 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr1_+_264741911 | 1.28 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr5_-_168734296 | 1.28 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr4_-_10995792 | 1.25 |
ENSRNOT00000078733
|
AABR07059243.1
|
|
| chr6_+_2216623 | 1.20 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr10_-_87753668 | 1.18 |
ENSRNOT00000066758
ENSRNOT00000047104 |
Krtap4-3
|
keratin associated protein 4-3 |
| chr15_-_26939911 | 1.18 |
ENSRNOT00000016683
|
Olr1630
|
olfactory receptor 1630 |
| chr4_+_1658278 | 1.14 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx1_Uncx_Prrx2_Shox2_Noto
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.4 | GO:0036233 | glycine import(GO:0036233) |
| 6.3 | 50.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 5.2 | 15.6 | GO:1990743 | protein sialylation(GO:1990743) |
| 4.3 | 55.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 3.8 | 15.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 3.3 | 10.0 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.9 | 5.7 | GO:0021550 | medulla oblongata development(GO:0021550) |
| 2.8 | 14.1 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.6 | 7.7 | GO:2001055 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 2.4 | 19.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 2.4 | 7.2 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 2.1 | 6.2 | GO:1903774 | multivesicular body assembly(GO:0036258) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.9 | 18.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 1.8 | 48.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 1.7 | 18.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.6 | 12.7 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 1.5 | 10.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.3 | 6.7 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 24.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 1.2 | 3.7 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 1.2 | 3.5 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 1.1 | 4.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 1.0 | 13.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.0 | 11.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.9 | 2.8 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.9 | 12.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.9 | 2.6 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.9 | 18.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.8 | 8.6 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.7 | 4.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 4.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.7 | 4.6 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.6 | 5.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.6 | 1.9 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.6 | 1.8 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.5 | 8.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 3.2 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.5 | 10.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.5 | 4.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 | 6.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.5 | 5.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 9.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 8.4 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.5 | 1.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.5 | 9.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.5 | 1.4 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.5 | 1.4 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.4 | 4.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 2.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.4 | 4.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 4.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.4 | 4.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 1.0 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.3 | 5.6 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.3 | 1.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.3 | 4.5 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 3.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.3 | 2.5 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 5.3 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.3 | 8.3 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.3 | 2.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 1.0 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 1.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 | 0.9 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 1.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.8 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 2.1 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 2.9 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 3.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 0.8 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 1.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 2.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 6.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 2.7 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 0.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 1.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.2 | 3.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 3.0 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 2.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 10.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 3.0 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 2.8 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.5 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 3.8 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 1.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 2.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.6 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.1 | 1.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 2.3 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 3.1 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.9 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 0.9 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 3.1 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 2.0 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.5 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 5.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 2.1 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.3 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.2 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.1 | 1.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 8.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.4 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 1.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.8 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 1.6 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 | 1.7 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 4.5 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.3 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 17.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.4 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 55.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 2.7 | 8.2 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 2.3 | 18.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.5 | 10.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.9 | 4.5 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 6.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.8 | 5.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.8 | 121.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.7 | 11.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 2.8 | GO:0042584 | secretory granule lumen(GO:0034774) chromaffin granule membrane(GO:0042584) |
| 0.7 | 6.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 3.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 10.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.4 | 21.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 18.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.3 | 2.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 0.9 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 7.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 2.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 4.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 12.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 2.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 16.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 12.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 9.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 6.1 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 3.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 6.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 4.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 7.8 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 20.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 12.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.7 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 5.1 | GO:0044445 | cytosolic part(GO:0044445) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 5.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 35.7 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 5.2 | 15.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 4.8 | 19.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 4.6 | 18.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 2.6 | 12.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 2.2 | 6.7 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 1.3 | 4.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 1.1 | 4.6 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.1 | 14.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 1.0 | 10.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.9 | 2.8 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.9 | 2.8 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.8 | 2.5 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.8 | 4.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.8 | 4.6 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.8 | 6.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.7 | 5.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.7 | 5.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 3.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.7 | 4.7 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.6 | 18.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.6 | 2.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.6 | 8.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 30.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.5 | 4.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.5 | 4.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.5 | 5.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 2.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.5 | 1.8 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 3.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 3.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.4 | 12.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 6.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 8.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 8.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 1.4 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 1.4 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 39.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 2.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 1.0 | GO:0003921 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.3 | 3.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 7.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 1.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 2.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 6.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 4.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.2 | 1.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 2.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.5 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.1 | 7.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 82.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 6.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.5 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 4.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 2.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 30.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 12.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 21.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 5.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 14.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.3 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.3 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 6.1 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 4.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 15.3 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 5.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 6.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 22.6 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 2.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 17.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 7.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 55.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.9 | 29.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.5 | 21.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.4 | 18.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 21.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 4.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 24.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 17.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 3.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 9.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 6.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 2.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 6.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 3.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 9.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 39.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 1.3 | 46.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 1.3 | 21.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.8 | 18.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.7 | 21.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.7 | 10.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.6 | 4.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.5 | 5.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 5.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.5 | 2.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 18.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 4.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.3 | 4.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 6.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 4.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 2.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 3.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 2.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 3.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 5.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 4.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 13.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.0 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |