Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
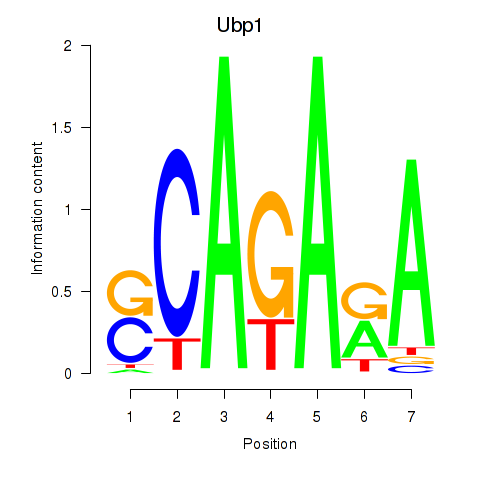
Results for Ubp1
Z-value: 0.88
Transcription factors associated with Ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ubp1
|
ENSRNOG00000009729 | upstream binding protein 1 (LBP-1a) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ubp1 | rn6_v1_chr8_+_122197027_122197027 | 0.26 | 2.7e-06 | Click! |
Activity profile of Ubp1 motif
Sorted Z-values of Ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_98457810 | 28.96 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr7_+_120153184 | 22.81 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr4_+_98370797 | 21.27 |
ENSRNOT00000031991
|
AABR07060872.1
|
|
| chr2_+_223029559 | 20.56 |
ENSRNOT00000045584
|
AABR07013111.1
|
|
| chr11_+_86092468 | 20.43 |
ENSRNOT00000057971
|
LOC100361706
|
lambda-chain C1-region-like |
| chrX_+_28539158 | 19.16 |
ENSRNOT00000073535
|
Tlr8
|
toll-like receptor 8 |
| chr3_+_19274273 | 18.19 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr4_+_106323089 | 17.38 |
ENSRNOT00000091402
|
AABR07061134.1
|
|
| chr1_-_227441442 | 17.12 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr3_-_2853272 | 16.62 |
ENSRNOT00000023022
|
Fcna
|
ficolin A |
| chr6_-_138508753 | 16.41 |
ENSRNOT00000006888
|
Ighm
|
immunoglobulin heavy constant mu |
| chr4_+_69403735 | 15.96 |
ENSRNOT00000082012
|
Trbv13-1
|
T cell receptor beta, variable 13-1 |
| chr4_+_14151343 | 15.90 |
ENSRNOT00000061687
ENSRNOT00000076573 ENSRNOT00000077219 ENSRNOT00000008319 |
Cd36
|
CD36 molecule |
| chr3_-_17081510 | 15.78 |
ENSRNOT00000063862
|
AABR07051562.1
|
|
| chr3_-_16752353 | 15.37 |
ENSRNOT00000046489
|
AABR07051548.1
|
|
| chr4_-_103258134 | 14.05 |
ENSRNOT00000086827
|
AABR07061052.1
|
|
| chr6_-_125723944 | 13.83 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr3_+_17107861 | 13.56 |
ENSRNOT00000043097
|
AABR07051563.1
|
|
| chr15_+_57241968 | 13.39 |
ENSRNOT00000082191
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr10_+_45258887 | 13.36 |
ENSRNOT00000048642
|
Btnl10
|
butyrophilin like 10 |
| chr4_+_70828894 | 12.73 |
ENSRNOT00000064892
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr6_-_142060032 | 12.62 |
ENSRNOT00000064717
|
AABR07065811.1
|
|
| chr5_+_173288447 | 12.24 |
ENSRNOT00000091205
ENSRNOT00000067252 |
Mxra8
|
matrix remodeling associated 8 |
| chr3_+_17139670 | 12.16 |
ENSRNOT00000073316
|
AABR07051564.1
|
|
| chr4_-_98305173 | 11.71 |
ENSRNOT00000010151
|
Il23r
|
interleukin 23 receptor |
| chr3_+_19174027 | 11.60 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chr2_-_60683451 | 11.06 |
ENSRNOT00000046215
|
Rai14
|
retinoic acid induced 14 |
| chr8_+_49282460 | 11.05 |
ENSRNOT00000021488
|
Cd3d
|
CD3d molecule |
| chr1_+_199555722 | 10.90 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
| chr5_+_144308611 | 10.80 |
ENSRNOT00000014388
|
Col8a2
|
collagen type VIII alpha 2 chain |
| chr20_-_32353677 | 10.65 |
ENSRNOT00000035355
|
Stox1
|
storkhead box 1 |
| chr8_+_55603968 | 10.58 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr3_+_20375699 | 10.58 |
ENSRNOT00000088492
|
AABR07051731.1
|
|
| chr6_-_143195143 | 10.53 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr6_-_125723732 | 10.43 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr6_-_142993147 | 10.36 |
ENSRNOT00000044381
|
LOC100359978
|
mCG1038839-like |
| chr1_-_73682247 | 10.23 |
ENSRNOT00000079498
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr3_+_17180411 | 10.20 |
ENSRNOT00000058260
|
AABR07051565.1
|
|
| chr7_-_18793289 | 10.11 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr8_-_125544427 | 10.02 |
ENSRNOT00000041548
ENSRNOT00000039904 ENSRNOT00000036642 |
Rbms3
|
RNA binding motif, single stranded interacting protein 3 |
| chr3_-_14643897 | 9.92 |
ENSRNOT00000082008
ENSRNOT00000025983 |
Ggta1
|
glycoprotein, alpha-galactosyltransferase 1 |
| chr3_-_110517163 | 9.70 |
ENSRNOT00000078037
|
Plcb2
|
phospholipase C, beta 2 |
| chr5_-_79691258 | 9.56 |
ENSRNOT00000072920
|
Tnfsf8
|
tumor necrosis factor superfamily member 8 |
| chr9_-_14668297 | 9.52 |
ENSRNOT00000042404
|
Treml2
|
triggering receptor expressed on myeloid cells-like 2 |
| chr6_-_141488290 | 9.31 |
ENSRNOT00000067336
|
AABR07065792.1
|
|
| chr12_+_19714324 | 9.30 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr19_-_10976396 | 9.25 |
ENSRNOT00000073239
|
Nlrc5
|
NLR family, CARD domain containing 5 |
| chr3_+_16817051 | 9.09 |
ENSRNOT00000071666
|
AABR07051550.1
|
|
| chr4_+_14109864 | 9.06 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr6_-_139997537 | 9.06 |
ENSRNOT00000073207
|
AABR07065740.1
|
|
| chr3_-_20695952 | 9.01 |
ENSRNOT00000072306
|
AABR07051746.1
|
|
| chr15_-_34603819 | 8.91 |
ENSRNOT00000067539
|
Cma1
|
chymase 1 |
| chr17_+_15814132 | 8.87 |
ENSRNOT00000032997
|
Susd3
|
sushi domain containing 3 |
| chr4_-_163227242 | 8.83 |
ENSRNOT00000091552
ENSRNOT00000077793 |
Clec7a
|
C-type lectin domain family 7, member A |
| chr3_-_20479999 | 8.81 |
ENSRNOT00000050573
|
AABR07051733.2
|
|
| chr12_-_20486276 | 8.80 |
ENSRNOT00000074057
|
LOC680910
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr12_+_25498198 | 8.68 |
ENSRNOT00000076916
|
Ncf1
|
neutrophil cytosolic factor 1 |
| chr10_-_56506446 | 8.53 |
ENSRNOT00000021357
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr3_+_19441604 | 8.48 |
ENSRNOT00000090581
|
AABR07051692.1
|
|
| chr4_+_90990088 | 8.39 |
ENSRNOT00000030320
|
Mmrn1
|
multimerin 1 |
| chr14_-_17225389 | 8.30 |
ENSRNOT00000052035
|
Art3
|
ADP-ribosyltransferase 3 |
| chr4_+_144192989 | 8.22 |
ENSRNOT00000007523
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr3_+_16495748 | 8.19 |
ENSRNOT00000045492
|
AABR07051533.1
|
|
| chrX_-_1346181 | 8.19 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chr12_+_48577905 | 8.17 |
ENSRNOT00000000888
|
Selplg
|
selectin P ligand |
| chr6_-_142372031 | 8.09 |
ENSRNOT00000063929
|
AABR07065814.3
|
|
| chr9_+_92618352 | 8.02 |
ENSRNOT00000034603
|
Sp140
|
SP140 nuclear body protein |
| chr4_-_70996395 | 8.00 |
ENSRNOT00000021485
|
Kel
|
Kell blood group, metallo-endopeptidase |
| chr6_-_140880070 | 7.99 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr8_-_63092009 | 7.97 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr1_+_47231093 | 7.91 |
ENSRNOT00000024967
|
Sytl3
|
synaptotagmin-like 3 |
| chr4_+_179398621 | 7.87 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr1_-_101236065 | 7.86 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr6_-_140642221 | 7.86 |
ENSRNOT00000081996
|
AABR07065772.2
|
|
| chr3_-_20419417 | 7.84 |
ENSRNOT00000077772
|
AABR07051733.3
|
|
| chr10_-_63491279 | 7.65 |
ENSRNOT00000082301
|
Tusc5
|
tumor suppressor candidate 5 |
| chr1_-_213650247 | 7.64 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr10_+_12046701 | 7.59 |
ENSRNOT00000011073
ENSRNOT00000084004 |
Mefv
|
MEFV, pyrin innate immunity regulator |
| chr8_+_117694605 | 7.57 |
ENSRNOT00000027994
|
Col7a1
|
collagen type VII alpha 1 chain |
| chr5_+_133865331 | 7.56 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr15_-_34612432 | 7.53 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr6_-_75874235 | 7.46 |
ENSRNOT00000009080
|
Baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr19_+_24456976 | 7.45 |
ENSRNOT00000004900
|
Ucp1
|
uncoupling protein 1 |
| chr2_-_149325913 | 7.45 |
ENSRNOT00000036690
|
Gpr171
|
G protein-coupled receptor 171 |
| chr4_+_101909389 | 7.43 |
ENSRNOT00000086458
|
AABR07060963.2
|
|
| chr7_-_29152442 | 7.42 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr14_-_18849258 | 7.40 |
ENSRNOT00000033406
|
Pf4
|
platelet factor 4 |
| chr17_-_54808483 | 7.32 |
ENSRNOT00000075683
|
AABR07028049.1
|
|
| chr11_+_17538063 | 7.23 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr16_-_48437223 | 7.17 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr8_-_133128290 | 7.15 |
ENSRNOT00000008783
|
Ccr1l1
|
chemokine (C-C motif) receptor 1-like 1 |
| chr7_+_144623555 | 7.13 |
ENSRNOT00000022217
|
Hoxc6
|
homeo box C6 |
| chr4_-_145390447 | 6.96 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chrX_-_105390580 | 6.94 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr6_-_141062581 | 6.94 |
ENSRNOT00000073446
|
AABR07065778.3
|
|
| chr6_-_143131118 | 6.90 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr1_+_168957460 | 6.87 |
ENSRNOT00000090745
|
LOC103694857
|
hemoglobin subunit beta-2 |
| chr10_+_45893018 | 6.79 |
ENSRNOT00000004280
ENSRNOT00000086710 |
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr4_-_156276304 | 6.78 |
ENSRNOT00000078725
|
Clec4e
|
C-type lectin domain family 4, member E |
| chr6_-_140485913 | 6.76 |
ENSRNOT00000048463
|
AABR07065768.1
|
|
| chr19_-_39267928 | 6.68 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr10_-_75171774 | 6.64 |
ENSRNOT00000011735
|
Epx
|
eosinophil peroxidase |
| chr15_+_32817343 | 6.64 |
ENSRNOT00000073853
|
AABR07017902.1
|
|
| chr12_-_21362205 | 6.52 |
ENSRNOT00000064787
|
LOC108348155
|
paired immunoglobulin-like type 2 receptor beta-2 |
| chr4_+_44597123 | 6.49 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr9_+_67774150 | 6.48 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr4_+_78694447 | 6.44 |
ENSRNOT00000011945
|
Gpnmb
|
glycoprotein nmb |
| chr12_-_21670269 | 6.43 |
ENSRNOT00000074863
|
LOC100910801
|
paired immunoglobulin-like type 2 receptor alpha-like |
| chr14_-_72122158 | 6.33 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr7_-_12424367 | 6.32 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr20_-_3374344 | 6.30 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr10_+_88326337 | 6.30 |
ENSRNOT00000022029
ENSRNOT00000076461 |
Fkbp10
|
FK506 binding protein 10 |
| chr6_-_138640187 | 6.28 |
ENSRNOT00000087983
|
AABR07065651.6
|
|
| chr15_+_57221292 | 6.26 |
ENSRNOT00000014502
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr4_+_69457472 | 6.26 |
ENSRNOT00000067597
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr4_+_14070553 | 6.25 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chrX_+_71174699 | 6.20 |
ENSRNOT00000076957
ENSRNOT00000090192 ENSRNOT00000040334 |
Med12
|
mediator complex subunit 12 |
| chr10_-_87067456 | 6.20 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr1_-_175796040 | 6.10 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr6_-_122239614 | 6.04 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr18_-_55859333 | 6.02 |
ENSRNOT00000025989
|
Myoz3
|
myozenin 3 |
| chr6_-_138772736 | 6.01 |
ENSRNOT00000071492
|
AABR07065651.1
|
|
| chr1_+_84304228 | 5.90 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr8_+_67753279 | 5.89 |
ENSRNOT00000009716
|
Calml4
|
calmodulin-like 4 |
| chr1_-_127337882 | 5.88 |
ENSRNOT00000085158
|
Aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr10_+_75087892 | 5.76 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr1_-_89045586 | 5.75 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr12_-_16126953 | 5.72 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr20_+_5040337 | 5.71 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr2_-_186515135 | 5.61 |
ENSRNOT00000077375
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chr3_-_166994286 | 5.60 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
| chr6_-_33738825 | 5.57 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr7_+_143060597 | 5.57 |
ENSRNOT00000087481
|
Krt7
|
keratin 7 |
| chrX_+_75382598 | 5.54 |
ENSRNOT00000033494
|
Uprt
|
uracil phosphoribosyltransferase homolog |
| chr2_+_208749996 | 5.54 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr6_-_141008427 | 5.52 |
ENSRNOT00000074472
|
AABR07065778.2
|
|
| chrX_-_157331204 | 5.50 |
ENSRNOT00000085201
|
Bgn
|
biglycan |
| chr17_-_79676499 | 5.45 |
ENSRNOT00000022711
|
Itga8
|
integrin subunit alpha 8 |
| chrX_-_138435391 | 5.43 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr6_+_93281408 | 5.41 |
ENSRNOT00000009610
|
Frmd6
|
FERM domain containing 6 |
| chr17_-_43614844 | 5.32 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr1_-_174495258 | 5.31 |
ENSRNOT00000052377
ENSRNOT00000086349 |
Scube2
|
signal peptide, CUB domain and EGF like domain containing 2 |
| chr4_-_165192647 | 5.31 |
ENSRNOT00000086461
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr19_+_61332351 | 5.29 |
ENSRNOT00000014492
|
Nrp1
|
neuropilin 1 |
| chr1_-_8751198 | 5.23 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr1_-_47307488 | 5.23 |
ENSRNOT00000090033
|
Ezr
|
ezrin |
| chr3_-_146470293 | 5.17 |
ENSRNOT00000009627
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr6_-_140418831 | 5.16 |
ENSRNOT00000086301
|
AABR07065768.2
|
|
| chr17_+_90696019 | 5.11 |
ENSRNOT00000003438
|
Gpr137b
|
G protein-coupled receptor 137B |
| chr4_-_78342863 | 5.11 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr3_-_119611136 | 5.03 |
ENSRNOT00000016157
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr17_-_9762813 | 4.97 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr1_+_81230612 | 4.96 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr12_+_19680712 | 4.93 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr6_-_140266507 | 4.92 |
ENSRNOT00000082865
|
AABR07065753.1
|
|
| chr6_+_24163026 | 4.92 |
ENSRNOT00000061284
|
Lbh
|
limb bud and heart development |
| chr3_+_112242270 | 4.92 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr1_-_169534896 | 4.92 |
ENSRNOT00000082645
|
Trim30c
|
tripartite motif-containing 30C |
| chr3_+_18787606 | 4.90 |
ENSRNOT00000090508
|
AABR07051658.1
|
|
| chr20_-_5227620 | 4.87 |
ENSRNOT00000086240
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr11_+_60383431 | 4.86 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr8_-_116045954 | 4.84 |
ENSRNOT00000020065
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr8_+_118392773 | 4.82 |
ENSRNOT00000066305
|
LOC100911725
|
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4-like |
| chr1_+_32221636 | 4.82 |
ENSRNOT00000022346
ENSRNOT00000089941 |
Slc6a18
|
solute carrier family 6 member 18 |
| chr13_+_89826254 | 4.81 |
ENSRNOT00000006141
|
F11r
|
F11 receptor |
| chr1_+_140601791 | 4.78 |
ENSRNOT00000091588
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr4_-_123713319 | 4.78 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr19_-_37448067 | 4.75 |
ENSRNOT00000092844
|
Zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr19_-_55300403 | 4.74 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr1_+_216293087 | 4.74 |
ENSRNOT00000027875
ENSRNOT00000087153 |
Kcnq1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr7_+_114997103 | 4.73 |
ENSRNOT00000010224
|
Ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr6_-_142299996 | 4.73 |
ENSRNOT00000086580
|
Ighv8-2
|
immunoglobulin heavy variable V8-2 |
| chr5_+_118541979 | 4.69 |
ENSRNOT00000013089
|
Efcab7
|
EF-hand calcium binding domain 7 |
| chr2_+_198388809 | 4.68 |
ENSRNOT00000083087
|
Hist2h2aa3
|
histone cluster 2, H2aa3 |
| chr4_-_149988295 | 4.62 |
ENSRNOT00000019649
|
Fxyd4
|
FXYD domain-containing ion transport regulator 4 |
| chr17_-_89163113 | 4.61 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr10_+_56193856 | 4.59 |
ENSRNOT00000080584
ENSRNOT00000091216 ENSRNOT00000083458 |
Tp53
|
tumor protein p53 |
| chr10_+_29165577 | 4.57 |
ENSRNOT00000078415
|
Ccnjl
|
cyclin J-like |
| chr10_-_4257868 | 4.56 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr13_+_50103189 | 4.56 |
ENSRNOT00000004078
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr13_+_88995316 | 4.56 |
ENSRNOT00000004117
|
Olfml2b
|
olfactomedin-like 2B |
| chr3_-_20050486 | 4.55 |
ENSRNOT00000088887
|
AABR07051718.1
|
|
| chr12_-_6879154 | 4.51 |
ENSRNOT00000001207
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr1_-_101086198 | 4.45 |
ENSRNOT00000027917
|
Rcn3
|
reticulocalbin 3 |
| chr12_-_9990284 | 4.43 |
ENSRNOT00000001264
|
Rasl11a
|
RAS-like family 11 member A |
| chr16_-_47207487 | 4.41 |
ENSRNOT00000059459
|
Dctd
|
dCMP deaminase |
| chr5_-_118541928 | 4.40 |
ENSRNOT00000012947
|
Itgb3bp
|
integrin subunit beta 3 binding protein |
| chr3_+_148541909 | 4.39 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr8_-_112884062 | 4.37 |
ENSRNOT00000016222
ENSRNOT00000085585 |
Acpp
|
acid phosphatase, prostate |
| chr11_-_64853692 | 4.34 |
ENSRNOT00000002089
|
Cd80
|
Cd80 molecule |
| chr20_+_1809078 | 4.34 |
ENSRNOT00000084853
|
Olr1739
|
olfactory receptor 1739 |
| chr10_-_89088993 | 4.34 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chr6_-_30187337 | 4.28 |
ENSRNOT00000074150
|
Itsn2
|
intersectin 2 |
| chr11_-_38088753 | 4.28 |
ENSRNOT00000002713
|
Tmprss2
|
transmembrane protease, serine 2 |
| chr5_+_104698040 | 4.27 |
ENSRNOT00000009127
|
Adamtsl1
|
ADAMTS-like 1 |
| chr1_+_157701089 | 4.24 |
ENSRNOT00000014271
|
Prcp
|
prolylcarboxypeptidase |
| chr8_-_132189388 | 4.22 |
ENSRNOT00000005817
|
Zdhhc3
|
zinc finger, DHHC-type containing 3 |
| chr6_+_50528823 | 4.21 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chr10_+_17261541 | 4.21 |
ENSRNOT00000005478
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr17_+_78879058 | 4.20 |
ENSRNOT00000022043
|
Olah
|
oleoyl-ACP hydrolase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 4.3 | 21.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 4.2 | 16.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 3.9 | 15.8 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 3.7 | 22.2 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 3.7 | 11.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 3.4 | 3.4 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 3.3 | 9.9 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 3.2 | 9.6 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 3.1 | 12.2 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 2.5 | 7.6 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 2.4 | 24.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 2.3 | 9.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 2.2 | 6.5 | GO:2000286 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) regulation of inward rectifier potassium channel activity(GO:1901979) positive regulation of gap junction assembly(GO:1903598) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 2.1 | 6.2 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 2.0 | 5.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 2.0 | 7.9 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 2.0 | 5.9 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.9 | 5.8 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 1.9 | 7.6 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 1.8 | 7.4 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.8 | 5.4 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 1.8 | 5.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.8 | 3.5 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 1.8 | 5.3 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.7 | 5.2 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 1.7 | 5.2 | GO:0019541 | acetate metabolic process(GO:0006083) propionate metabolic process(GO:0019541) |
| 1.7 | 6.8 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) positive regulation of T-helper 17 type immune response(GO:2000318) |
| 1.7 | 5.0 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 1.6 | 4.9 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 1.6 | 4.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.5 | 3.1 | GO:0032752 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.5 | 10.6 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 1.5 | 4.6 | GO:0045763 | regulation of arginine metabolic process(GO:0000821) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.5 | 4.4 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 1.5 | 4.4 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 1.4 | 5.7 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.4 | 4.2 | GO:0002353 | kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 1.4 | 4.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.4 | 4.1 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.3 | 4.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.3 | 8.0 | GO:0031133 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 1.3 | 6.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 1.3 | 3.8 | GO:0042891 | antibiotic transport(GO:0042891) |
| 1.2 | 9.9 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 1.2 | 4.9 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 1.2 | 6.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 1.2 | 3.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 4.6 | GO:1904024 | negative regulation of fermentation(GO:1901003) negative regulation of NAD metabolic process(GO:1902689) negative regulation of glucose catabolic process to lactate via pyruvate(GO:1904024) |
| 1.1 | 4.5 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.1 | 6.7 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.1 | 5.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.1 | 4.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.1 | 23.5 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 1.1 | 5.3 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 1.1 | 3.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 1.0 | 5.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.9 | 2.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.9 | 5.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.9 | 2.6 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.8 | 4.2 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.8 | 11.7 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 0.8 | 3.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.8 | 4.9 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.8 | 11.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.8 | 9.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.8 | 5.5 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.8 | 4.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.8 | 11.0 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.8 | 2.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.7 | 7.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 2.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.7 | 2.9 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.7 | 7.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.7 | 9.9 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.7 | 8.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.7 | 5.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 | 10.8 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.6 | 2.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.6 | 2.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.6 | 3.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 7.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.6 | 6.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.6 | 4.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.6 | 3.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.6 | 4.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.6 | 6.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.6 | 1.8 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.6 | 2.3 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.6 | 7.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.5 | 4.9 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.5 | 2.6 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.5 | 1.5 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.5 | 3.6 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.5 | 2.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 5.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.5 | 1.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.5 | 2.9 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.5 | 1.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.5 | 3.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 2.4 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.5 | 3.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.5 | 4.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.5 | 1.4 | GO:0002333 | transitional stage B cell differentiation(GO:0002332) transitional one stage B cell differentiation(GO:0002333) positive regulation of actin filament binding(GO:1904531) positive regulation of actin binding(GO:1904618) |
| 0.5 | 1.9 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.5 | 7.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.5 | 2.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.5 | 6.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 1.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.4 | 3.5 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 3.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 7.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.4 | 0.4 | GO:0043132 | NAD transport(GO:0043132) |
| 0.4 | 5.8 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.4 | 6.9 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.4 | 1.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 2.4 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.4 | 2.0 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.4 | 9.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.4 | 3.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.4 | 1.9 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 5.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.4 | 4.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.8 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.4 | 2.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.4 | 2.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 1.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.4 | 3.2 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 0.4 | 2.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.3 | 9.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.3 | 1.0 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.3 | 1.6 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.0 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.3 | 2.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.3 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.5 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.3 | 1.8 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.3 | 1.5 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) |
| 0.3 | 1.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.3 | 11.6 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.3 | 1.9 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.3 | 6.6 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.3 | 0.8 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 | 2.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.3 | 9.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.3 | 2.4 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.3 | 1.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.3 | 6.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 3.8 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.3 | 1.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 1.0 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.2 | 0.7 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.2 | 1.9 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 2.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 1.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 | 2.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 2.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 0.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.4 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 3.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 4.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 0.6 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.2 | 9.3 | GO:0035904 | aorta development(GO:0035904) |
| 0.2 | 5.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 0.2 | GO:0036334 | epidermal stem cell homeostasis(GO:0036334) |
| 0.2 | 2.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.2 | 1.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.2 | 1.4 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.2 | 1.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 2.8 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 7.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.2 | 2.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 1.3 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.2 | 4.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 2.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 2.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 6.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 6.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.4 | GO:0071221 | response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 2.4 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 3.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 1.0 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 2.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 5.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 1.5 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 3.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.2 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.1 | 3.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 2.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.9 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 3.6 | GO:0006220 | pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.1 | 3.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.5 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 1.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.9 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.4 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 3.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 1.1 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.6 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 1.2 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 0.6 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 1.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.6 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 10.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 2.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.1 | 3.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 4.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.6 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 4.3 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 4.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.7 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.8 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.0 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 3.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 5.8 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.1 | 0.5 | GO:0071680 | response to indole-3-methanol(GO:0071680) |
| 0.1 | 2.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 3.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 3.5 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 3.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 7.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 0.3 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 3.3 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 2.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 2.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 7.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 2.0 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.5 | GO:0060028 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) convergent extension involved in axis elongation(GO:0060028) convergent extension involved in organogenesis(GO:0060029) |
| 0.1 | 2.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 | 2.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 5.0 | GO:1903052 | positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) |
| 0.1 | 1.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 3.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.7 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.1 | 1.6 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 2.0 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 1.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 2.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 2.0 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 4.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 2.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 3.3 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 1.9 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 2.8 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.3 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 2.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.7 | GO:0042110 | T cell activation(GO:0042110) T cell aggregation(GO:0070489) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 24.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 3.2 | 9.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.4 | 16.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.4 | 11.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.3 | 5.2 | GO:0036398 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 1.2 | 8.6 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 1.1 | 4.2 | GO:0005607 | laminin-1 complex(GO:0005606) laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 1.0 | 9.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.9 | 11.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.8 | 19.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.7 | 6.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.7 | 2.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.7 | 7.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.6 | 5.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.6 | 5.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.6 | 6.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.6 | 7.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 2.9 | GO:0043293 | apoptosome(GO:0043293) |
| 0.5 | 20.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.5 | 3.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 1.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.5 | 3.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 2.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 4.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 2.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.4 | 3.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.4 | 36.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 3.6 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 5.8 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 7.4 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.3 | 1.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.3 | 3.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 4.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 2.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 4.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 7.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 2.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.3 | 0.8 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 1.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 17.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 4.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 2.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 34.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 2.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 5.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 2.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 15.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 1.4 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 17.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.2 | 15.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 0.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.2 | 10.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 2.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 6.8 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 7.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 3.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 3.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 5.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.1 | GO:0097452 | CRD-mediated mRNA stability complex(GO:0070937) GAIT complex(GO:0097452) |
| 0.1 | 4.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) site of double-strand break(GO:0035861) |
| 0.1 | 11.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 3.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 7.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 29.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 5.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 7.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 5.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 4.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 14.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 5.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 5.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 3.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 5.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 5.8 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.1 | 1.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 4.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 9.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 9.5 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 6.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 9.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.5 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 22.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 3.8 | 22.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.9 | 11.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 2.5 | 9.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 2.2 | 6.7 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 2.0 | 25.5 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 1.9 | 7.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.8 | 11.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.6 | 4.8 | GO:0030977 | taurine binding(GO:0030977) |
| 1.5 | 7.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.4 | 13.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.4 | 4.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.3 | 5.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 1.3 | 5.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.3 | 19.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 1.3 | 3.8 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 1.2 | 31.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 1.2 | 9.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 1.2 | 4.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 1.1 | 5.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 1.0 | 22.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 1.0 | 5.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 1.0 | 5.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.0 | 2.9 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.0 | 4.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.9 | 5.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.9 | 3.5 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.8 | 6.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 3.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.8 | 3.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.8 | 3.8 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.7 | 7.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.7 | 5.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.7 | 7.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.7 | 5.3 | GO:0038085 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 0.7 | 5.9 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.6 | 3.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 1.9 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.6 | 3.7 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.6 | 7.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.6 | 4.8 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.6 | 3.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 7.4 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.6 | 3.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.6 | 3.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.5 | 2.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 2.6 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.5 | 2.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.5 | 4.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.5 | 3.5 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 1.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.5 | 2.0 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.5 | 6.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 3.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.5 | 4.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.5 | 6.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 4.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.4 | 4.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 4.4 | GO:0033265 | choline binding(GO:0033265) |
| 0.4 | 2.5 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 2.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.4 | 2.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 4.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 2.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.4 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.4 | 5.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.4 | 1.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 0.4 | 7.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 2.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 5.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 8.0 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 3.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 16.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 7.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 16.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 0.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.3 | 2.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 3.0 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.3 | 3.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 4.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 3.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 1.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.3 | 2.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 11.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.3 | 1.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 4.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.3 | 21.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.3 | 2.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 2.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 14.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 1.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 0.8 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 3.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 2.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 1.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.3 | 1.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 0.7 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.2 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 3.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 3.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.7 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 20.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 7.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 2.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 3.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 8.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 16.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.2 | 1.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.8 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.2 | 0.6 | GO:0003921 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.2 | 2.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 1.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 4.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 4.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.2 | 1.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 4.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 1.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.6 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.2 | 1.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.8 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 2.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 3.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 3.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 7.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 2.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 5.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 2.7 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 4.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.8 | GO:0004445 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.6 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 1.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 12.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.4 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.2 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.1 | 2.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 3.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.1 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 8.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 7.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 5.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 6.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.3 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.6 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 1.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 6.0 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 7.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.9 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 2.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 4.7 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 9.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 3.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 4.7 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 10.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 1.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 17.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 17.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 9.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 5.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 7.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.5 | 17.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.5 | 15.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 6.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 14.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.4 | 8.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 4.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.3 | 16.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 11.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 9.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 15.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 4.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 5.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 3.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 32.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 5.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 6.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 2.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 1.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 5.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 3.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 3.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 4.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 6.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 4.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 9.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 19.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.4 | 11.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.2 | 14.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.0 | 9.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 5.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 5.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 28.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 5.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 6.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.4 | 5.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 6.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 2.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.4 | 28.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 2.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 6.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 19.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 4.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 12.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 28.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 4.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.3 | 10.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 5.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 3.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 3.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 3.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.3 | 6.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 6.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 6.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 1.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 4.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 3.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 11.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 5.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 5.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 7.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 4.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 2.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 6.5 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.2 | 0.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.2 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 2.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 5.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 4.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.8 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.1 | 6.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |