Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
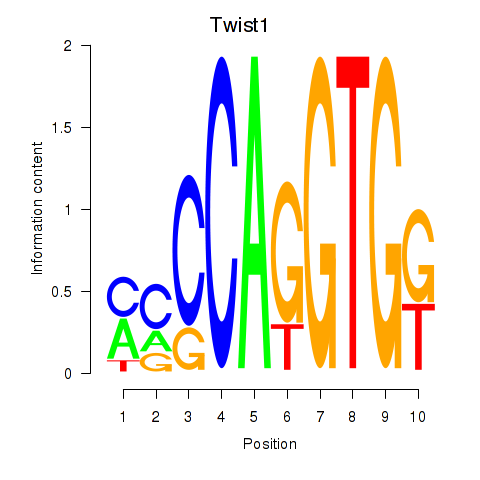
Results for Twist1
Z-value: 0.45
Transcription factors associated with Twist1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Twist1
|
ENSRNOG00000011101 | twist family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | rn6_v1_chr6_+_53401109_53401109 | -0.15 | 6.4e-03 | Click! |
Activity profile of Twist1 motif
Sorted Z-values of Twist1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_69268336 | 29.20 |
ENSRNOT00000018042
|
Prss3b
|
protease, serine, 3B |
| chr19_-_37907714 | 15.90 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr4_-_69196430 | 13.55 |
ENSRNOT00000017673
|
LOC312273
|
Trypsin V-A |
| chr20_-_5123073 | 11.29 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr1_+_100470722 | 10.89 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr13_+_89826254 | 7.86 |
ENSRNOT00000006141
|
F11r
|
F11 receptor |
| chr3_+_5624506 | 7.54 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr3_-_170955399 | 7.19 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr1_+_263448633 | 5.33 |
ENSRNOT00000064510
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr10_+_91187593 | 4.88 |
ENSRNOT00000004163
|
Acbd4
|
acyl-CoA binding domain containing 4 |
| chr10_-_90999506 | 4.80 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr12_+_17416327 | 4.42 |
ENSRNOT00000089590
ENSRNOT00000092186 |
Adap1
|
ArfGAP with dual PH domains 1 |
| chr5_-_169212170 | 4.37 |
ENSRNOT00000013385
|
Tas1r1
|
taste 1 receptor member 1 |
| chr10_+_58776792 | 4.37 |
ENSRNOT00000019574
|
Txndc17
|
thioredoxin domain containing 17 |
| chr1_+_80383050 | 4.07 |
ENSRNOT00000023451
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr15_-_33428942 | 4.03 |
ENSRNOT00000019618
ENSRNOT00000092160 |
Slc7a8
|
solute carrier family 7 member 8 |
| chr1_-_221431713 | 4.02 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr19_+_58823814 | 3.89 |
ENSRNOT00000027058
ENSRNOT00000088626 |
Kcnk1
|
potassium two pore domain channel subfamily K member 1 |
| chr1_-_32272476 | 3.84 |
ENSRNOT00000022683
|
Tert
|
telomerase reverse transcriptase |
| chr9_+_27343853 | 3.83 |
ENSRNOT00000072794
|
Tmem14a
|
transmembrane protein 14A |
| chr20_-_35449280 | 3.62 |
ENSRNOT00000057422
|
Man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr19_+_50045020 | 3.49 |
ENSRNOT00000090165
|
Plcg2
|
phospholipase C, gamma 2 |
| chr8_-_48672732 | 3.07 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr3_+_35271786 | 3.00 |
ENSRNOT00000065989
|
Lypd6b
|
LY6/PLAUR domain containing 6B |
| chr10_+_15088935 | 2.98 |
ENSRNOT00000030273
|
Gng13
|
G protein subunit gamma 13 |
| chr10_+_13000090 | 2.63 |
ENSRNOT00000004845
|
Cldn6
|
claudin 6 |
| chr3_-_51297852 | 2.61 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr1_+_81474553 | 2.50 |
ENSRNOT00000083493
|
Phldb3
|
pleckstrin homology-like domain, family B, member 3 |
| chr20_+_14101659 | 2.20 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr11_+_68198709 | 2.14 |
ENSRNOT00000003048
|
Dirc2
|
disrupted in renal carcinoma 2 |
| chr1_+_101214593 | 1.82 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr9_-_17827032 | 1.81 |
ENSRNOT00000026929
|
Slc35b2
|
solute carrier family 35 member B2 |
| chrX_-_157286936 | 1.59 |
ENSRNOT00000078100
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr2_-_186245342 | 1.44 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr12_-_38691075 | 1.43 |
ENSRNOT00000084192
|
Bcl7a
|
BCL tumor suppressor 7A |
| chr1_+_127802978 | 1.37 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr1_-_67134827 | 1.36 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr20_+_3830164 | 1.31 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr2_-_186245163 | 1.24 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr7_-_97759852 | 1.14 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr5_+_62071346 | 1.14 |
ENSRNOT00000012478
|
Anp32b
|
acidic nuclear phosphoprotein 32 family member B |
| chr4_+_140703619 | 1.09 |
ENSRNOT00000009563
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chrX_+_33599671 | 1.09 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr18_-_63488027 | 1.09 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr8_-_55171718 | 0.93 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr7_-_12274803 | 0.78 |
ENSRNOT00000049146
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr19_+_55246926 | 0.63 |
ENSRNOT00000017358
|
Il17c
|
interleukin 17C |
| chr3_-_130114770 | 0.60 |
ENSRNOT00000010638
|
Jag1
|
jagged 1 |
| chr15_-_3435888 | 0.51 |
ENSRNOT00000016709
|
Adk
|
adenosine kinase |
| chr5_+_39251337 | 0.45 |
ENSRNOT00000010073
|
Ndufaf4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr1_-_242152834 | 0.43 |
ENSRNOT00000071614
ENSRNOT00000020412 |
Fxn
|
frataxin |
| chr1_-_89309379 | 0.43 |
ENSRNOT00000028533
|
Ffar1
|
free fatty acid receptor 1 |
| chr2_-_30791221 | 0.41 |
ENSRNOT00000082846
|
Ccnb1
|
cyclin B1 |
| chr20_+_3830376 | 0.31 |
ENSRNOT00000085547
|
Col11a2
|
collagen type XI alpha 2 chain |
| chr10_+_75032365 | 0.25 |
ENSRNOT00000010448
|
Supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chrX_+_159513800 | 0.23 |
ENSRNOT00000065636
|
Htatsf1
|
HIV-1 Tat specific factor 1 |
| chr4_-_184096806 | 0.23 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr10_+_87774552 | 0.21 |
ENSRNOT00000044342
|
Krtap9-1
|
keratin associated protein 9-1 |
| chr20_-_2103864 | 0.20 |
ENSRNOT00000001014
|
Rnf39
|
ring finger protein 39 |
| chr18_-_29340403 | 0.09 |
ENSRNOT00000025157
|
Hbegf
|
heparin-binding EGF-like growth factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Twist1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 3.6 | 10.9 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 2.4 | 7.2 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 1.3 | 3.8 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.9 | 7.5 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.8 | 4.8 | GO:1904714 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.7 | 4.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.6 | 3.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.5 | 3.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.4 | 7.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.4 | 1.1 | GO:1903972 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) regulation of prolactin signaling pathway(GO:1902211) regulation of interleukin-4-mediated signaling pathway(GO:1902214) regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.3 | 3.5 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.3 | 4.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.3 | 1.8 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.3 | 1.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 42.7 | GO:0007586 | digestion(GO:0007586) |
| 0.2 | 1.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 0.6 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.2 | 2.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 3.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.5 | GO:0009216 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) AMP salvage(GO:0044209) adenosine biosynthetic process(GO:0046086) |
| 0.1 | 0.4 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 2.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.4 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) cellular response to iron(III) ion(GO:0071283) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.1 | 1.8 | GO:0048368 | notochord development(GO:0030903) lateral mesoderm development(GO:0048368) |
| 0.1 | 4.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 2.9 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 0.6 | GO:1900017 | neutrophil differentiation(GO:0030223) positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 1.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 4.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 3.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.2 | 4.8 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.8 | 7.9 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.6 | 1.8 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.5 | 1.6 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 1.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 3.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 4.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 8.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 3.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 45.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.8 | 3.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.6 | 7.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.5 | 4.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.5 | 3.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.4 | 10.9 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.4 | 1.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.3 | 4.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 3.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 59.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 3.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 4.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.8 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.2 | 3.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 4.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 1.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 4.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 11.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 4.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 8.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) metallochaperone activity(GO:0016530) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 7.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 1.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 7.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 7.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 8.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 3.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 4.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 4.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 2.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 4.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 3.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |