Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
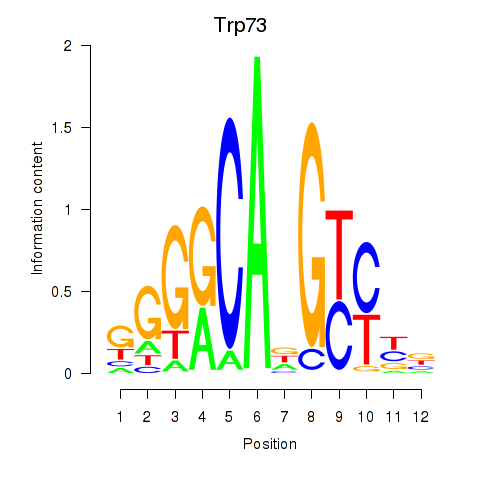
Results for Trp73
Z-value: 0.53
Transcription factors associated with Trp73
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tp73 | rn6_v1_chr5_-_171415354_171415354 | 0.03 | 6.2e-01 | Click! |
Activity profile of Trp73 motif
Sorted Z-values of Trp73 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_162385575 | 20.86 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr8_-_62948720 | 14.28 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr14_+_7618022 | 10.23 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr16_-_69132584 | 10.11 |
ENSRNOT00000017776
|
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr13_+_100980574 | 9.70 |
ENSRNOT00000067005
ENSRNOT00000004649 |
Capn8
|
calpain 8 |
| chr2_-_88314254 | 9.13 |
ENSRNOT00000038546
|
Car13
|
carbonic anhydrase 13 |
| chr11_-_73678984 | 8.93 |
ENSRNOT00000002355
|
Fam43a
|
family with sequence similarity 43, member A |
| chr3_+_110734105 | 8.64 |
ENSRNOT00000073587
|
Chst14
|
carbohydrate sulfotransferase 14 |
| chr9_-_61690956 | 8.12 |
ENSRNOT00000066589
|
Hspd1
|
heat shock protein family D member 1 |
| chr10_+_88326337 | 7.52 |
ENSRNOT00000022029
ENSRNOT00000076461 |
Fkbp10
|
FK506 binding protein 10 |
| chr10_+_88326080 | 7.15 |
ENSRNOT00000076475
|
Fkbp10
|
FK506 binding protein 10 |
| chr11_-_81334120 | 6.69 |
ENSRNOT00000089988
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr3_+_45683993 | 6.56 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr5_+_119903507 | 6.37 |
ENSRNOT00000033933
|
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr3_+_110723481 | 5.88 |
ENSRNOT00000013878
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr15_+_120372 | 5.66 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr10_+_103395511 | 5.66 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr1_-_213987053 | 5.36 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr8_-_115388266 | 4.72 |
ENSRNOT00000018122
|
Tex264
|
testis expressed 264 |
| chr10_+_15241590 | 4.04 |
ENSRNOT00000037372
ENSRNOT00000037381 |
Mettl26
|
methyltransferase like 26 |
| chr20_+_5933595 | 3.62 |
ENSRNOT00000000618
|
Mapk14
|
mitogen activated protein kinase 14 |
| chr5_-_166924136 | 3.19 |
ENSRNOT00000085251
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr18_-_24823837 | 3.15 |
ENSRNOT00000021405
ENSRNOT00000090390 |
Myo7b
|
myosin VIIb |
| chr7_-_47928674 | 2.76 |
ENSRNOT00000038144
|
Mettl25
|
methyltransferase like 25 |
| chr10_-_15590220 | 2.62 |
ENSRNOT00000048977
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr9_+_16647922 | 2.60 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr8_+_117455262 | 2.60 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr15_+_34552410 | 2.52 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr3_+_11921715 | 2.43 |
ENSRNOT00000021689
|
Fam129b
|
family with sequence similarity 129, member B |
| chrX_-_13116743 | 2.34 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr1_+_48008683 | 2.34 |
ENSRNOT00000018934
|
Acat2l1
|
acetyl-CoA acetyltransferase 2-like 1 |
| chr11_+_60383431 | 2.17 |
ENSRNOT00000093295
|
Cd200
|
Cd200 molecule |
| chr9_-_16647458 | 2.10 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr20_-_5455632 | 1.97 |
ENSRNOT00000000552
|
Wdr46
|
WD repeat domain 46 |
| chr1_-_87777668 | 1.97 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr2_-_250981623 | 1.93 |
ENSRNOT00000018435
|
Clca2
|
chloride channel accessory 2 |
| chr8_+_106503504 | 1.84 |
ENSRNOT00000018755
|
Rbp2
|
retinol binding protein 2 |
| chr10_+_52334240 | 1.76 |
ENSRNOT00000005518
|
Zfp18
|
zinc finger protein 18 |
| chr1_+_77541359 | 1.73 |
ENSRNOT00000049028
|
LOC103690015
|
40S ribosomal protein S19-like |
| chr5_+_59008933 | 1.67 |
ENSRNOT00000023060
|
Car9
|
carbonic anhydrase 9 |
| chr3_-_153114520 | 1.66 |
ENSRNOT00000008254
|
Dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr5_+_59080765 | 1.63 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr9_+_16647598 | 1.56 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr12_+_47218969 | 1.19 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr4_-_129846642 | 1.17 |
ENSRNOT00000010717
|
Frmd4b
|
FERM domain containing 4B |
| chr5_-_153924896 | 1.14 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr18_+_57011575 | 1.14 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr10_-_78219793 | 1.12 |
ENSRNOT00000003369
|
Tom1l1
|
target of myb1 like 1 membrane trafficking protein |
| chr10_+_85978691 | 1.02 |
ENSRNOT00000006359
|
Rpl19
|
ribosomal protein L19 |
| chr10_+_55478298 | 0.84 |
ENSRNOT00000038454
|
Rnf222
|
ring finger protein 222 |
| chr3_-_72113680 | 0.78 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr20_-_43108198 | 0.74 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr1_-_167588630 | 0.23 |
ENSRNOT00000050707
|
Olr39
|
olfactory receptor 39 |
| chr6_-_132797675 | 0.11 |
ENSRNOT00000006128
|
Wars
|
tryptophanyl-tRNA synthetase |
| chr7_+_47928060 | 0.04 |
ENSRNOT00000005740
|
Ccdc59
|
coiled-coil domain containing 59 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp73
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 2.7 | 8.1 | GO:0045041 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) protein import into mitochondrial intermembrane space(GO:0045041) |
| 1.7 | 6.7 | GO:0072312 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 1.4 | 5.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.1 | 8.6 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.9 | 3.6 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.8 | 20.9 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.6 | 2.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.6 | 3.2 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.4 | 5.9 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 14.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 10.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 2.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.3 | 1.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 6.6 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 0.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 2.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 2.4 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 1.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 1.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 9.7 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 2.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 10.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 6.4 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 2.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.0 | GO:0097421 | liver regeneration(GO:0097421) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 2.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.7 | 5.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.4 | 1.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 6.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 7.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 9.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 6.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.0 | 5.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 15.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 10.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 13.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.4 | 8.6 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 1.1 | 14.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.9 | 2.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.9 | 2.6 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.8 | 10.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 6.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 3.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) NFAT protein binding(GO:0051525) |
| 0.4 | 9.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.7 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 1.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 1.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 10.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 5.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 7.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 1.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 16.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.3 | GO:0016747 | transferase activity, transferring acyl groups other than amino-acyl groups(GO:0016747) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.6 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 8.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 8.6 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 4.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 6.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |