Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
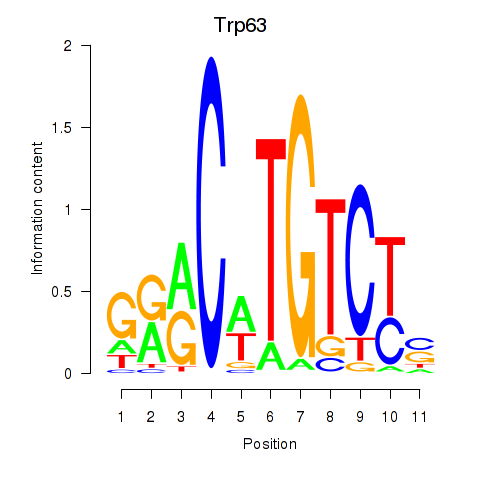
Results for Trp63
Z-value: 0.35
Transcription factors associated with Trp63
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tp63 | rn6_v1_chr11_-_78341349_78341349 | 0.22 | 5.6e-05 | Click! |
Activity profile of Trp63 motif
Sorted Z-values of Trp63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_80807181 | 17.65 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
| chr10_-_56465393 | 8.96 |
ENSRNOT00000056860
|
Tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr16_-_69132584 | 7.64 |
ENSRNOT00000017776
|
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr16_-_50501716 | 7.02 |
ENSRNOT00000067486
|
Fat1
|
FAT atypical cadherin 1 |
| chr12_-_23661009 | 6.98 |
ENSRNOT00000059451
|
Upk3bl
|
uroplakin 3B-like |
| chr16_-_50501921 | 6.76 |
ENSRNOT00000081023
|
Fat1
|
FAT atypical cadherin 1 |
| chr17_-_90627101 | 5.82 |
ENSRNOT00000003349
|
Nid1
|
nidogen 1 |
| chr2_-_219097619 | 5.13 |
ENSRNOT00000078806
|
Vcam1
|
vascular cell adhesion molecule 1 |
| chr6_-_77848434 | 4.63 |
ENSRNOT00000034342
|
Slc25a21
|
solute carrier family 25 member 21 |
| chr1_-_59347472 | 4.21 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr1_-_213987053 | 3.98 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr1_+_68436917 | 3.81 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr4_+_62220736 | 3.47 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr4_+_62221011 | 3.45 |
ENSRNOT00000041264
|
Cald1
|
caldesmon 1 |
| chr5_+_58667885 | 3.43 |
ENSRNOT00000064755
|
Unc13b
|
unc-13 homolog B |
| chr10_-_10881844 | 3.09 |
ENSRNOT00000087118
ENSRNOT00000004416 |
Mgrn1
|
mahogunin ring finger 1 |
| chr14_-_114583122 | 2.84 |
ENSRNOT00000084595
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_-_144269486 | 2.80 |
ENSRNOT00000090051
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr20_+_11489456 | 2.72 |
ENSRNOT00000001632
|
Lrrc3
|
leucine rich repeat containing 3 |
| chr5_+_135354700 | 2.58 |
ENSRNOT00000021850
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr7_-_31784192 | 2.44 |
ENSRNOT00000010869
|
Apaf1
|
apoptotic peptidase activating factor 1 |
| chr1_-_214423881 | 2.21 |
ENSRNOT00000025290
|
Pidd1
|
p53-induced death domain protein 1 |
| chr11_-_62128044 | 1.98 |
ENSRNOT00000093141
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr7_-_2961873 | 1.89 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr7_-_143497108 | 1.73 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr10_-_89374516 | 1.60 |
ENSRNOT00000028084
|
Vat1
|
vesicle amine transport 1 |
| chr18_+_60392376 | 1.38 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr13_+_48679774 | 1.35 |
ENSRNOT00000075674
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr9_-_16807966 | 1.32 |
ENSRNOT00000073160
|
Rps19
|
ribosomal protein S19 |
| chr15_+_34552410 | 1.09 |
ENSRNOT00000027802
|
Khnyn
|
KH and NYN domain containing |
| chr1_+_81750928 | 1.09 |
ENSRNOT00000027246
|
NEWGENE_68440
|
ribosomal protein S19 |
| chr15_-_33537146 | 0.67 |
ENSRNOT00000019992
|
Ppp1r3e
|
protein phosphatase 1, regulatory subunit 3E |
| chr16_+_61130755 | 0.58 |
ENSRNOT00000042609
|
AABR07026048.1
|
|
| chr10_-_64657089 | 0.42 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr3_+_75848904 | 0.29 |
ENSRNOT00000042996
|
Olr583
|
olfactory receptor 583 |
| chr19_+_22699808 | 0.29 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr2_-_250981623 | 0.24 |
ENSRNOT00000018435
|
Clca2
|
chloride channel accessory 2 |
| chr5_-_38923095 | 0.20 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr6_-_132797675 | 0.16 |
ENSRNOT00000006128
|
Wars
|
tryptophanyl-tRNA synthetase |
| chr13_+_78805347 | 0.06 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp63
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 2.5 | 7.6 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 1.4 | 4.2 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.5 | 5.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.5 | 3.4 | GO:0099525 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.4 | 5.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 3.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 | 1.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 1.3 | GO:0019042 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.3 | 1.3 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 13.8 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.3 | 2.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 1.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.3 | 2.8 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.2 | 3.8 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 | 2.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 0.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 9.0 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 1.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 2.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 2.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 6.9 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 2.7 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.3 | 17.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 1.2 | 6.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 2.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.4 | 3.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 4.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 2.8 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.2 | 2.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 5.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 13.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 9.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 3.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 17.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.7 | 5.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.6 | 4.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.5 | 5.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.4 | 3.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 2.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 3.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 9.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 6.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 2.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 13.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 3.1 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 2.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 2.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.0 | 17.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 6.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 5.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 4.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |