Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Trp53
Z-value: 1.30
Transcription factors associated with Trp53
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tp53 | rn6_v1_chr10_+_56187020_56187020 | 0.80 | 8.7e-73 | Click! |
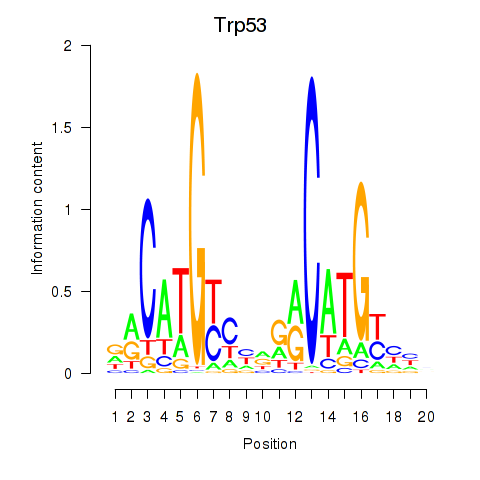
Activity profile of Trp53 motif
Sorted Z-values of Trp53 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_82369493 | 50.82 |
ENSRNOT00000003733
|
Sell
|
selectin L |
| chr20_+_5509059 | 47.46 |
ENSRNOT00000065349
|
Kifc1
|
kinesin family member C1 |
| chr17_+_44738643 | 45.95 |
ENSRNOT00000087643
|
LOC100910554
|
histone H2A type 1-like |
| chr4_-_170841187 | 45.61 |
ENSRNOT00000007485
|
Art4
|
ADP-ribosyltransferase 4 |
| chr4_+_78382287 | 44.57 |
ENSRNOT00000084927
|
Gimap5
|
GTPase, IMAP family member 5 |
| chr3_-_92749121 | 40.85 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr13_+_27465930 | 38.67 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr5_+_149056078 | 37.56 |
ENSRNOT00000083028
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr13_+_26903052 | 37.03 |
ENSRNOT00000003625
|
Serpinb5
|
serpin family B member 5 |
| chr19_+_19395655 | 36.20 |
ENSRNOT00000019130
|
Snx20
|
sorting nexin 20 |
| chr13_+_48427038 | 35.30 |
ENSRNOT00000009241
|
Ctse
|
cathepsin E |
| chr3_-_55451798 | 32.59 |
ENSRNOT00000008837
|
Spc25
|
SPC25, NDC80 kinetochore complex component |
| chr3_+_18706988 | 32.43 |
ENSRNOT00000074650
|
AABR07051652.1
|
|
| chr1_-_214423881 | 30.12 |
ENSRNOT00000025290
|
Pidd1
|
p53-induced death domain protein 1 |
| chr20_+_7788084 | 27.65 |
ENSRNOT00000000597
|
Def6
|
DEF6 guanine nucleotide exchange factor |
| chr16_+_74752655 | 27.58 |
ENSRNOT00000029266
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr15_+_35002406 | 27.32 |
ENSRNOT00000091167
|
Mcpt1l4
|
mast cell protease 1-like 4 |
| chr8_+_47674321 | 27.10 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr17_-_43798383 | 25.99 |
ENSRNOT00000075069
|
LOC684828
|
similar to Histone H1.2 (H1 VAR.1) (H1c) |
| chr1_-_227175096 | 25.94 |
ENSRNOT00000054811
|
AABR07006259.1
|
|
| chr12_+_1903165 | 25.05 |
ENSRNOT00000083947
|
AABR07034956.1
|
|
| chr10_-_107424710 | 24.94 |
ENSRNOT00000004320
|
Lgals3bp
|
galectin 3 binding protein |
| chr5_-_137104993 | 24.45 |
ENSRNOT00000027271
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr5_-_153924896 | 24.08 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr7_+_126736732 | 24.05 |
ENSRNOT00000022012
|
Gtse1
|
G-2 and S-phase expressed 1 |
| chr17_+_44520537 | 23.68 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr20_+_3987230 | 23.36 |
ENSRNOT00000088615
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr10_-_34242985 | 22.77 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr17_-_44520240 | 22.53 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr1_-_89329418 | 22.53 |
ENSRNOT00000033155
|
Cd22
|
CD22 molecule |
| chr9_-_15410943 | 22.46 |
ENSRNOT00000074217
|
Ccnd3
|
cyclin D3 |
| chr2_+_96439286 | 21.97 |
ENSRNOT00000016091
|
Il7
|
interleukin 7 |
| chr13_-_74520634 | 21.87 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_+_77079402 | 21.14 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr15_+_55126953 | 21.09 |
ENSRNOT00000021430
|
Lpar6
|
lysophosphatidic acid receptor 6 |
| chr17_+_77224112 | 20.34 |
ENSRNOT00000024178
|
Mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr16_-_14360555 | 20.15 |
ENSRNOT00000065460
|
LOC290595
|
hypothetical gene supported by AF152002 |
| chr11_+_64522130 | 19.78 |
ENSRNOT00000004315
|
Upk1b
|
uroplakin 1B |
| chr11_-_60547201 | 19.50 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr3_-_119611136 | 19.50 |
ENSRNOT00000016157
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr13_-_27158628 | 18.35 |
ENSRNOT00000003475
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr20_-_32139789 | 17.86 |
ENSRNOT00000078140
|
Srgn
|
serglycin |
| chr4_+_162437089 | 17.71 |
ENSRNOT00000038801
|
Clec2d2
|
C-type lectin domain family 2 member D2 |
| chr16_-_20383337 | 17.49 |
ENSRNOT00000025977
|
Il12rb1
|
interleukin 12 receptor subunit beta 1 |
| chr4_-_157263890 | 17.33 |
ENSRNOT00000065416
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_-_64853692 | 17.02 |
ENSRNOT00000002089
|
Cd80
|
Cd80 molecule |
| chr1_-_162385575 | 17.01 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr4_-_22307453 | 16.70 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr8_+_85489553 | 16.62 |
ENSRNOT00000082158
|
Rpl12-ps1
|
ribosomal protein L12, pseudogene 1 |
| chr16_+_20110148 | 16.61 |
ENSRNOT00000080146
ENSRNOT00000025312 |
Jak3
|
Janus kinase 3 |
| chr4_+_162292305 | 16.17 |
ENSRNOT00000010098
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr9_-_121931564 | 16.06 |
ENSRNOT00000056243
|
Tyms
|
thymidylate synthetase |
| chr18_+_62174670 | 15.98 |
ENSRNOT00000025362
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr8_+_71125414 | 15.81 |
ENSRNOT00000021265
|
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chrX_-_104984341 | 15.79 |
ENSRNOT00000077996
|
Xkrx
|
XK related, X-linked |
| chr2_-_250981623 | 15.51 |
ENSRNOT00000018435
|
Clca2
|
chloride channel accessory 2 |
| chrX_+_28539158 | 15.49 |
ENSRNOT00000073535
|
Tlr8
|
toll-like receptor 8 |
| chr11_-_60546997 | 15.37 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr3_-_156340913 | 15.36 |
ENSRNOT00000021452
|
Mafb
|
MAF bZIP transcription factor B |
| chr10_+_107469377 | 15.32 |
ENSRNOT00000087404
|
C1qtnf1
|
C1q and tumor necrosis factor related protein 1 |
| chr17_-_44527801 | 15.24 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr10_-_63176463 | 15.11 |
ENSRNOT00000004717
|
Slc6a4
|
solute carrier family 6 member 4 |
| chr17_+_44528125 | 15.03 |
ENSRNOT00000084538
|
LOC680322
|
similar to Histone H2A type 1 |
| chr12_+_38160464 | 15.02 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr11_+_84745904 | 14.71 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr1_+_84573496 | 14.67 |
ENSRNOT00000057158
|
RGD1565183
|
similar to ribosomal protein L28 |
| chr13_+_47454591 | 14.41 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr15_-_34603819 | 14.35 |
ENSRNOT00000067539
|
Cma1
|
chymase 1 |
| chr10_-_91027731 | 14.29 |
ENSRNOT00000084128
ENSRNOT00000093704 |
Kif18b
|
kinesin family member 18B |
| chr1_+_75233703 | 13.83 |
ENSRNOT00000064178
|
LOC100911727
|
DNA ligase 1-like |
| chr3_+_177164359 | 13.53 |
ENSRNOT00000066837
|
RGD1561282
|
RGD1561282 |
| chr8_+_72743426 | 13.43 |
ENSRNOT00000072573
|
Rps27l
|
ribosomal protein S27-like |
| chr10_-_90415070 | 13.28 |
ENSRNOT00000055179
|
Itga2b
|
integrin subunit alpha 2b |
| chr6_+_24163026 | 13.14 |
ENSRNOT00000061284
|
Lbh
|
limb bud and heart development |
| chr16_-_85306366 | 13.11 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr5_+_135442031 | 12.71 |
ENSRNOT00000090852
ENSRNOT00000034464 |
Ccdc17
|
coiled-coil domain containing 17 |
| chr1_+_77541359 | 12.69 |
ENSRNOT00000049028
|
LOC103690015
|
40S ribosomal protein S19-like |
| chr14_-_81023682 | 12.46 |
ENSRNOT00000081570
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr16_+_2743823 | 12.44 |
ENSRNOT00000087873
|
Arhgef3
|
Rho guanine nucleotide exchange factor 3 |
| chr7_-_143852119 | 12.33 |
ENSRNOT00000016801
|
Rarg
|
retinoic acid receptor, gamma |
| chr9_-_78693028 | 12.22 |
ENSRNOT00000036359
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr3_+_21764377 | 12.21 |
ENSRNOT00000063938
|
Gpr21
|
G protein-coupled receptor 21 |
| chr8_+_66296883 | 12.21 |
ENSRNOT00000018467
|
Tle3
|
transducin-like enhancer of split 3 |
| chr3_+_2643610 | 11.71 |
ENSRNOT00000037169
|
Fut7
|
fucosyltransferase 7 |
| chr5_+_162808646 | 11.59 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr2_-_206293599 | 11.54 |
ENSRNOT00000026213
|
Dclre1b
|
DNA cross-link repair 1B |
| chr1_+_88078350 | 11.38 |
ENSRNOT00000048677
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr5_+_171478555 | 11.21 |
ENSRNOT00000091417
ENSRNOT00000000169 |
LOC100911486
|
multiple epidermal growth factor-like domains protein 6-like |
| chr20_-_6257604 | 11.20 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chr18_-_4371899 | 10.77 |
ENSRNOT00000051207
|
LOC108349606
|
60S ribosomal protein L7a-like |
| chr18_-_25314047 | 10.72 |
ENSRNOT00000079778
|
AABR07031674.4
|
|
| chr1_-_213987053 | 10.57 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr8_+_128781395 | 10.57 |
ENSRNOT00000029766
|
Ccr8
|
C-C motif chemokine receptor 8 |
| chr2_+_30685840 | 10.45 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr2_-_189765415 | 10.42 |
ENSRNOT00000020815
|
Slc27a3
|
solute carrier family 27 member 3 |
| chr7_-_2588843 | 10.30 |
ENSRNOT00000088619
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr7_-_123892429 | 10.25 |
ENSRNOT00000037681
|
Nfam1
|
NFAT activating protein with ITAM motif 1 |
| chrX_-_139916883 | 10.23 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr8_+_49378644 | 10.17 |
ENSRNOT00000007588
|
Jaml
|
junction adhesion molecule like |
| chr10_+_11100917 | 9.94 |
ENSRNOT00000006067
|
Coro7
|
coronin 7 |
| chr12_-_13133219 | 9.88 |
ENSRNOT00000092292
|
Daglb
|
diacylglycerol lipase, beta |
| chr17_-_8429338 | 9.86 |
ENSRNOT00000016390
|
Tgfbi
|
transforming growth factor, beta induced |
| chr7_+_13938302 | 9.63 |
ENSRNOT00000009643
|
Casp14
|
caspase 14 |
| chr10_-_14061703 | 9.61 |
ENSRNOT00000017917
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr17_-_78499881 | 9.57 |
ENSRNOT00000079260
|
Fam107b
|
family with sequence similarity 107, member B |
| chr5_-_155258392 | 9.46 |
ENSRNOT00000017065
|
C1qc
|
complement C1q C chain |
| chr6_-_141365198 | 9.42 |
ENSRNOT00000040523
|
AABR07065789.2
|
|
| chr2_-_206293275 | 9.23 |
ENSRNOT00000080702
|
Dclre1b
|
DNA cross-link repair 1B |
| chrX_-_31138675 | 9.23 |
ENSRNOT00000004456
|
Fancb
|
Fanconi anemia, complementation group B |
| chr6_+_80188943 | 9.13 |
ENSRNOT00000059335
|
Mia2
|
melanoma inhibitory activity 2 |
| chr2_-_95106157 | 9.10 |
ENSRNOT00000041334
|
LOC499573
|
LRRGT00056 |
| chr10_+_45289741 | 8.44 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr4_-_148446303 | 8.38 |
ENSRNOT00000017633
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr10_+_83104622 | 8.28 |
ENSRNOT00000072972
|
AABR07030375.2
|
|
| chr4_+_100407658 | 7.97 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr7_-_117068332 | 7.91 |
ENSRNOT00000082433
|
Fam83h
|
family with sequence similarity 83, member H |
| chr3_-_125607735 | 7.69 |
ENSRNOT00000028901
|
Fermt1
|
fermitin family member 1 |
| chr20_+_11365697 | 7.68 |
ENSRNOT00000001611
|
Aire
|
autoimmune regulator |
| chr3_-_72895740 | 7.57 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr8_-_83693472 | 7.51 |
ENSRNOT00000015947
|
Fam83b
|
family with sequence similarity 83, member B |
| chr13_+_48644604 | 7.47 |
ENSRNOT00000084259
ENSRNOT00000073669 |
Rab29
|
RAB29, member RAS oncogene family |
| chr1_+_103172987 | 7.42 |
ENSRNOT00000018688
|
Tmem86a
|
transmembrane protein 86A |
| chr10_+_107502695 | 7.40 |
ENSRNOT00000038088
|
Engase
|
endo-beta-N-acetylglucosaminidase |
| chr8_-_120446455 | 7.35 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr9_+_95241609 | 7.33 |
ENSRNOT00000032634
ENSRNOT00000077416 |
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr10_-_12990405 | 7.32 |
ENSRNOT00000078647
|
Thoc6
|
THO complex 6 |
| chrX_-_71616997 | 7.15 |
ENSRNOT00000004406
|
Cxcr3
|
C-X-C motif chemokine receptor 3 |
| chr7_-_71048383 | 7.02 |
ENSRNOT00000005693
|
Gpr182
|
G protein-coupled receptor 182 |
| chr16_+_20020444 | 7.01 |
ENSRNOT00000024827
|
Pgls
|
6-phosphogluconolactonase |
| chr19_-_29802083 | 6.96 |
ENSRNOT00000024755
|
AC123471.1
|
|
| chr4_+_6282278 | 6.91 |
ENSRNOT00000010349
|
Kmt2c
|
lysine methyltransferase 2C |
| chr20_-_5166252 | 6.85 |
ENSRNOT00000001138
|
Aif1
|
allograft inflammatory factor 1 |
| chr11_+_86715981 | 6.75 |
ENSRNOT00000050269
|
Comt
|
catechol-O-methyltransferase |
| chr8_+_106503504 | 6.59 |
ENSRNOT00000018755
|
Rbp2
|
retinol binding protein 2 |
| chr1_+_239412884 | 6.56 |
ENSRNOT00000067384
|
Tmem2
|
transmembrane protein 2 |
| chr3_+_151285249 | 6.49 |
ENSRNOT00000055254
|
Procr
|
protein C receptor |
| chr6_-_76270457 | 6.32 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chr1_+_226573105 | 6.21 |
ENSRNOT00000028074
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr13_+_112031594 | 6.19 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr19_+_15339152 | 6.11 |
ENSRNOT00000060929
|
Ces1a
|
carboxylesterase 1A |
| chr14_+_83343330 | 6.11 |
ENSRNOT00000089198
|
Pisd
|
phosphatidylserine decarboxylase |
| chr13_-_89343868 | 6.10 |
ENSRNOT00000058497
ENSRNOT00000035400 |
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr17_+_78879058 | 5.98 |
ENSRNOT00000022043
|
Olah
|
oleoyl-ACP hydrolase |
| chr7_-_117679219 | 5.95 |
ENSRNOT00000071522
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr6_+_10912383 | 5.92 |
ENSRNOT00000061747
ENSRNOT00000086247 |
Ttc7a
|
tetratricopeptide repeat domain 7A |
| chr7_+_118507224 | 5.90 |
ENSRNOT00000075451
|
LOC108348142
|
60S ribosomal protein L8 |
| chr1_-_151106802 | 5.75 |
ENSRNOT00000021971
|
Tyr
|
tyrosinase |
| chr8_+_33514042 | 5.74 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr11_+_53140599 | 5.73 |
ENSRNOT00000083438
|
Bbx
|
BBX, HMG-box containing |
| chr2_+_154604832 | 5.69 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr7_-_2986935 | 5.69 |
ENSRNOT00000081125
ENSRNOT00000006578 |
Pa2g4
|
proliferation-associated 2G4 |
| chr6_-_111176918 | 5.56 |
ENSRNOT00000016242
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chrX_-_38026774 | 5.43 |
ENSRNOT00000074898
|
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr10_-_56511583 | 5.32 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr7_-_28711761 | 5.20 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chrX_+_22302485 | 5.17 |
ENSRNOT00000082902
|
Kdm5c
|
lysine demethylase 5C |
| chr8_-_108801570 | 5.11 |
ENSRNOT00000091593
|
Nck1
|
NCK adaptor protein 1 |
| chr5_-_137014375 | 5.04 |
ENSRNOT00000066165
ENSRNOT00000091866 |
Kdm4a
|
lysine demethylase 4A |
| chr4_+_102290338 | 5.02 |
ENSRNOT00000011067
|
AABR07060992.1
|
|
| chr1_+_201429771 | 5.01 |
ENSRNOT00000027836
|
Plekha1
|
pleckstrin homology domain containing A1 |
| chr1_-_129776276 | 4.98 |
ENSRNOT00000051402
|
Arrdc4
|
arrestin domain containing 4 |
| chr10_+_84955864 | 4.92 |
ENSRNOT00000038572
|
Sp6
|
Sp6 transcription factor |
| chr10_+_31561895 | 4.87 |
ENSRNOT00000048485
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr12_-_30180115 | 4.86 |
ENSRNOT00000001202
|
Crcp
|
CGRP receptor component |
| chr5_+_133865331 | 4.84 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr9_-_100306829 | 4.84 |
ENSRNOT00000038563
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr20_-_3439983 | 4.68 |
ENSRNOT00000080822
ENSRNOT00000001099 |
Ier3
|
immediate early response 3 |
| chr14_+_107349414 | 4.64 |
ENSRNOT00000012255
|
LOC690096
|
similar to ribosomal protein L28 |
| chr17_-_84488480 | 4.53 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr7_-_140401686 | 4.42 |
ENSRNOT00000083955
|
Fkbp11
|
FK506 binding protein 11 |
| chr20_+_46707362 | 4.41 |
ENSRNOT00000071045
|
AABR07045411.1
|
|
| chr1_+_140584380 | 4.39 |
ENSRNOT00000064438
|
Aen
|
apoptosis enhancing nuclease |
| chr6_+_80218880 | 4.36 |
ENSRNOT00000006995
|
Mia2
|
melanoma inhibitory activity 2 |
| chr5_-_152473868 | 4.32 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr5_-_113621134 | 4.27 |
ENSRNOT00000037206
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr20_+_1889652 | 4.25 |
ENSRNOT00000068508
ENSRNOT00000000996 |
Olr1746
|
olfactory receptor 1746 |
| chr17_-_17947777 | 4.25 |
ENSRNOT00000036876
|
Rnf144b
|
ring finger protein 144B |
| chr9_+_17216495 | 4.25 |
ENSRNOT00000026331
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr17_+_90802393 | 4.22 |
ENSRNOT00000003535
|
Edaradd
|
EDAR-associated death domain |
| chr4_-_150616895 | 4.22 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chrX_-_84167717 | 4.13 |
ENSRNOT00000006415
|
Pof1b
|
premature ovarian failure 1B |
| chr3_-_64766472 | 4.02 |
ENSRNOT00000037684
|
Cwc22
|
CWC22 spliceosome associated protein homolog |
| chr7_+_72985495 | 3.91 |
ENSRNOT00000008361
|
Matn2
|
matrilin 2 |
| chr3_-_122006803 | 3.88 |
ENSRNOT00000089660
|
RGD1566226
|
similar to hypothetical protein F830045P16 |
| chr10_+_25890943 | 3.85 |
ENSRNOT00000086814
|
Nudcd2
|
NudC domain containing 2 |
| chr3_-_162080192 | 3.84 |
ENSRNOT00000025820
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr8_-_119012671 | 3.75 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr4_-_117758135 | 3.70 |
ENSRNOT00000087750
|
Nat8f4
|
N-acetyltransferase 8 (GCN5-related) family member 4 |
| chr13_-_111849653 | 3.67 |
ENSRNOT00000006395
|
Diexf
|
digestive organ expansion factor homolog (zebrafish) |
| chr1_-_198638944 | 3.57 |
ENSRNOT00000023285
|
Cd2bp2
|
Cd2 (cytoplasmic tail) binding protein 2 |
| chr6_+_143901280 | 3.54 |
ENSRNOT00000091884
ENSRNOT00000005648 |
Zfp386
|
zinc finger protein 386 (Kruppel-like) |
| chr1_+_175839108 | 3.54 |
ENSRNOT00000023741
|
Ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr5_+_159735008 | 3.54 |
ENSRNOT00000064310
|
Rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr6_+_26686033 | 3.51 |
ENSRNOT00000081167
|
Slc5a6
|
solute carrier family 5 member 6 |
| chr1_-_266914093 | 3.50 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr19_+_11450760 | 3.50 |
ENSRNOT00000026297
|
Nudt21
|
nudix hydrolase 21 |
| chr1_-_164307084 | 3.49 |
ENSRNOT00000086091
|
Serpinh1
|
serpin family H member 1 |
| chr5_-_101138427 | 3.44 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr19_+_49637016 | 3.41 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chr8_+_130749838 | 3.36 |
ENSRNOT00000079273
|
Snrk
|
SNF related kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Trp53
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 40.9 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 7.8 | 23.4 | GO:0046967 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 7.4 | 44.6 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 6.0 | 17.9 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 5.6 | 50.8 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 5.5 | 16.6 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 5.5 | 21.9 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 5.4 | 16.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 5.3 | 16.0 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 5.2 | 15.5 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 4.8 | 9.6 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 4.8 | 14.3 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 4.3 | 17.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 4.2 | 16.7 | GO:1905235 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 4.2 | 20.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 4.1 | 12.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 4.1 | 24.4 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 3.9 | 19.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 3.9 | 34.9 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 3.8 | 15.4 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 3.8 | 7.7 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 3.8 | 15.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 3.2 | 26.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 3.2 | 9.6 | GO:0070268 | cornification(GO:0070268) |
| 3.2 | 15.8 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 3.1 | 9.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 3.0 | 15.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 3.0 | 9.0 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 2.6 | 13.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 2.6 | 10.2 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 2.5 | 15.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 2.4 | 12.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 2.3 | 13.8 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 2.3 | 6.8 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 2.3 | 6.8 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 2.2 | 13.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 2.2 | 35.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 2.2 | 13.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 2.1 | 8.4 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 2.1 | 12.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 2.1 | 31.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 2.0 | 5.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.0 | 23.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 1.9 | 5.7 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 1.9 | 5.6 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.8 | 12.5 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 1.7 | 5.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 1.7 | 10.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 1.7 | 5.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.7 | 27.0 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 1.6 | 4.9 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 1.6 | 42.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 1.6 | 9.5 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 1.5 | 6.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 1.5 | 4.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.5 | 14.7 | GO:0002467 | germinal center formation(GO:0002467) |
| 1.4 | 11.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.4 | 4.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.3 | 17.5 | GO:0002827 | positive regulation of T-helper 1 type immune response(GO:0002827) |
| 1.3 | 11.7 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 1.3 | 2.6 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 1.3 | 47.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 1.3 | 3.8 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 1.3 | 6.3 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 1.2 | 3.5 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 1.2 | 3.5 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 1.1 | 10.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.1 | 17.0 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 1.1 | 21.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 1.1 | 15.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 1.1 | 3.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 1.0 | 22.0 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 1.0 | 15.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.9 | 37.0 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.9 | 7.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.9 | 20.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.9 | 6.0 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.8 | 0.8 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.8 | 6.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.8 | 7.3 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.8 | 2.3 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 0.8 | 2.3 | GO:0002086 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) diaphragm contraction(GO:0002086) |
| 0.7 | 69.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.7 | 2.1 | GO:0002001 | renin secretion into blood stream(GO:0002001) |
| 0.7 | 3.5 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.7 | 4.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.7 | 7.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.7 | 20.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.7 | 39.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.7 | 17.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.6 | 5.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.6 | 5.0 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.6 | 22.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.6 | 21.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.6 | 9.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.6 | 9.9 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.6 | 1.7 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.5 | 4.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.5 | 1.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 2.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.5 | 7.2 | GO:1900118 | T cell chemotaxis(GO:0010818) negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.5 | 2.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 4.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.5 | 3.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 32.6 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.4 | 12.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 4.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.4 | 7.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.4 | 6.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 5.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 8.0 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.4 | 3.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.4 | 2.6 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 1.8 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.4 | 31.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 4.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 5.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.3 | 27.1 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.3 | 12.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.3 | 5.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 13.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 4.9 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.2 | 2.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 6.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 1.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 7.9 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.2 | 3.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 3.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 10.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 0.6 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.2 | 11.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.2 | 2.4 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 2.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 4.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 16.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 7.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.7 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 1.5 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.1 | 2.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 8.9 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 1.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 7.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 24.6 | GO:0016485 | protein processing(GO:0016485) |
| 0.1 | 4.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 5.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 9.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 24.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 3.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 3.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 9.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 2.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 2.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.9 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 3.9 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 1.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 9.9 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 2.0 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.3 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 6.2 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 1.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.6 | 40.9 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 6.5 | 32.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 3.3 | 23.4 | GO:0042825 | TAP complex(GO:0042825) |
| 2.4 | 19.5 | GO:0000796 | condensin complex(GO:0000796) |
| 2.2 | 17.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.0 | 12.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 2.0 | 20.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 1.8 | 14.3 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 1.5 | 7.5 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 1.4 | 141.8 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 1.3 | 4.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 1.3 | 16.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.1 | 8.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.9 | 51.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.9 | 2.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.8 | 8.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.8 | 47.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.7 | 10.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.7 | 7.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.7 | 6.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 6.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.7 | 13.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.7 | 26.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.7 | 17.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.6 | 9.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 6.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 3.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.5 | 3.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 38.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.5 | 145.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 22.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.4 | 1.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 26.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.4 | 26.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 1.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.3 | 6.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 6.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 2.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 4.9 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 29.1 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 2.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 5.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 25.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 20.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 14.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 9.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 4.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 15.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 17.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 4.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 98.5 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.2 | 7.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 14.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.2 | 8.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 17.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 8.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 2.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 23.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 8.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 28.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 7.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 69.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 2.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 18.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 17.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 18.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 6.1 | 24.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 5.7 | 45.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 5.3 | 21.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 5.0 | 15.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 4.4 | 17.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 4.2 | 41.7 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 4.1 | 12.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 3.9 | 19.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 3.3 | 16.7 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 3.2 | 9.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 3.0 | 20.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 2.8 | 8.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 2.6 | 15.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 2.4 | 14.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 2.4 | 4.9 | GO:0001635 | calcitonin gene-related peptide receptor activity(GO:0001635) |
| 2.4 | 14.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 2.3 | 7.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 2.2 | 13.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 2.2 | 50.8 | GO:0051861 | glycolipid binding(GO:0051861) |
| 2.1 | 21.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 2.1 | 12.5 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 1.6 | 16.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.6 | 7.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.5 | 6.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 1.5 | 13.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.5 | 10.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.4 | 5.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 1.4 | 5.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.4 | 35.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 1.3 | 27.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.3 | 17.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.2 | 11.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.2 | 10.4 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 1.1 | 5.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.0 | 12.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.0 | 7.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 1.0 | 5.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.9 | 7.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.9 | 9.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.9 | 2.6 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.9 | 33.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.8 | 97.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.8 | 22.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.8 | 2.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.7 | 6.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.7 | 61.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.7 | 10.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.7 | 15.0 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.6 | 2.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.6 | 5.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.6 | 6.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 4.4 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.5 | 11.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 10.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 24.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.5 | 2.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 3.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.4 | 1.7 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.4 | 48.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.4 | 6.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.4 | 3.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 9.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.4 | 2.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 12.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 6.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 13.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 25.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 1.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 7.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 3.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 6.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 2.3 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.3 | 17.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 4.5 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 5.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 3.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 7.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 7.7 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 3.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 5.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 6.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 15.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) single-stranded RNA binding(GO:0003727) |
| 0.2 | 116.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 2.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 37.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 3.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 3.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 5.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 7.3 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.1 | 21.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 3.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 10.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 45.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 12.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 3.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 6.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.4 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 7.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 1.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 16.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 5.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 3.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 3.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 10.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 3.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 4.3 | GO:0008134 | transcription factor binding(GO:0008134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 91.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.5 | 16.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 1.3 | 81.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.8 | 20.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.7 | 22.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.7 | 17.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.7 | 52.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.5 | 20.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 19.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 17.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 12.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.4 | 6.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 23.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.4 | 54.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 9.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 67.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 15.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 16.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 7.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 9.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 6.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 6.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 5.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 4.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 33.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 3.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 41.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 2.6 | 38.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 1.8 | 21.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 1.7 | 17.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 1.7 | 15.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.5 | 47.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 1.5 | 37.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 1.3 | 23.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 1.2 | 21.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.2 | 56.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.9 | 9.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.8 | 16.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.8 | 13.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.8 | 20.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.7 | 17.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.7 | 26.6 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.7 | 16.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.7 | 22.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.7 | 12.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.6 | 6.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.6 | 10.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.6 | 9.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 9.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.5 | 12.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 5.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 32.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.4 | 7.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.4 | 9.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.4 | 17.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 4.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 5.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 12.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 5.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 2.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 17.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 9.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 23.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 6.6 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 5.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 8.7 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |