Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
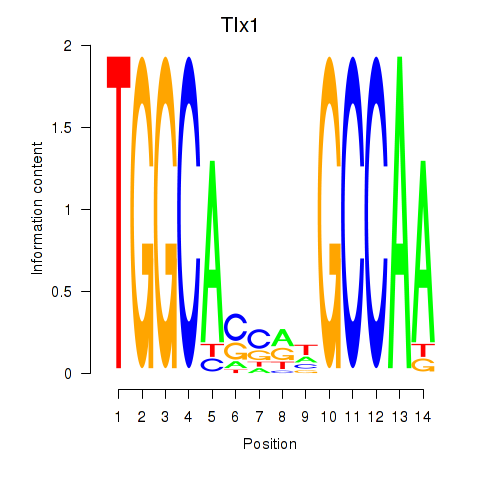
Results for Tlx1
Z-value: 0.82
Transcription factors associated with Tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tlx1
|
ENSRNOG00000016120 | T-cell leukemia, homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tlx1 | rn6_v1_chr1_+_264893162_264893162 | -0.73 | 5.4e-55 | Click! |
Activity profile of Tlx1 motif
Sorted Z-values of Tlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_85386231 | 45.16 |
ENSRNOT00000015316
|
Inmt
|
indolethylamine N-methyltransferase |
| chr13_-_80738634 | 26.05 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_134526089 | 25.84 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr13_+_113692131 | 23.78 |
ENSRNOT00000057160
|
Cd34
|
CD34 molecule |
| chr16_-_74122889 | 22.56 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chr13_+_113691932 | 20.43 |
ENSRNOT00000084709
|
Cd34
|
CD34 molecule |
| chr1_-_162385575 | 19.58 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr18_+_68408890 | 16.66 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr12_-_6740714 | 16.07 |
ENSRNOT00000001205
|
Medag
|
mesenteric estrogen-dependent adipogenesis |
| chr2_-_26011429 | 14.75 |
ENSRNOT00000065143
|
Arhgef28
|
Rho guanine nucleotide exchange factor 28 |
| chr5_-_93244202 | 14.12 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr13_-_80379929 | 13.93 |
ENSRNOT00000067653
ENSRNOT00000075938 |
Dnm3
|
dynamin 3 |
| chr4_-_147163467 | 13.23 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr1_+_91433030 | 12.78 |
ENSRNOT00000015205
|
Slc7a10
|
solute carrier family 7 member 10 |
| chr1_+_154579949 | 12.58 |
ENSRNOT00000045048
|
Sytl2
|
synaptotagmin-like 2 |
| chr10_+_84147836 | 12.27 |
ENSRNOT00000010527
|
Hoxb6
|
homeo box B6 |
| chr4_-_17594598 | 12.11 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr1_-_261986759 | 11.91 |
ENSRNOT00000021257
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr5_-_147412705 | 11.63 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr1_-_156354353 | 11.55 |
ENSRNOT00000042163
|
Sytl2
|
synaptotagmin-like 2 |
| chr6_-_137084739 | 11.23 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chr5_-_101166651 | 11.14 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr1_-_100669684 | 11.11 |
ENSRNOT00000091760
|
Myh14
|
myosin heavy chain 14 |
| chr4_+_62019970 | 11.04 |
ENSRNOT00000013133
|
LOC100910708
|
aldose reductase-related protein 1-like |
| chr20_-_9855443 | 10.82 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr13_+_47454591 | 10.22 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr3_+_149668102 | 10.03 |
ENSRNOT00000055342
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr14_+_84306466 | 10.00 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr7_+_143749221 | 9.70 |
ENSRNOT00000014807
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr2_-_195908037 | 9.64 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr3_-_160853650 | 9.41 |
ENSRNOT00000018844
|
Matn4
|
matrilin 4 |
| chr16_+_6078122 | 9.24 |
ENSRNOT00000021407
|
Chdh
|
choline dehydrogenase |
| chr13_+_112031594 | 9.09 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr3_+_35271786 | 8.92 |
ENSRNOT00000065989
|
Lypd6b
|
LY6/PLAUR domain containing 6B |
| chr3_-_175601127 | 8.83 |
ENSRNOT00000081226
|
Lama5
|
laminin subunit alpha 5 |
| chr2_-_195888216 | 8.76 |
ENSRNOT00000056436
|
Tuft1
|
tuftelin 1 |
| chr3_-_110556808 | 8.75 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr20_+_5008508 | 8.67 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr1_+_214778496 | 8.53 |
ENSRNOT00000028967
|
Muc5b
|
mucin 5B, oligomeric mucus/gel-forming |
| chr3_-_105512939 | 8.50 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr18_-_58499836 | 8.46 |
ENSRNOT00000065693
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr7_-_143016040 | 8.35 |
ENSRNOT00000029697
|
Krt80
|
keratin 80 |
| chr4_-_117111653 | 8.34 |
ENSRNOT00000057537
|
Sfxn5
|
sideroflexin 5 |
| chr4_-_114853868 | 8.26 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr3_+_112428395 | 8.19 |
ENSRNOT00000079109
ENSRNOT00000048141 |
Stard9
|
StAR-related lipid transfer domain containing 9 |
| chr6_+_119519714 | 8.10 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_6083083 | 7.84 |
ENSRNOT00000072411
|
Sulf1
|
sulfatase 1 |
| chr16_+_54291251 | 7.65 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr5_-_126395668 | 7.62 |
ENSRNOT00000010396
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr2_+_95320283 | 7.47 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr14_+_84237520 | 7.43 |
ENSRNOT00000083580
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr4_-_82177420 | 7.41 |
ENSRNOT00000074452
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr20_-_13142856 | 7.38 |
ENSRNOT00000001743
|
S100b
|
S100 calcium binding protein B |
| chr3_+_110975923 | 7.38 |
ENSRNOT00000016458
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr4_+_85662892 | 7.14 |
ENSRNOT00000016175
ENSRNOT00000043851 ENSRNOT00000035722 ENSRNOT00000046192 |
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor type 1 |
| chr8_+_116776494 | 7.08 |
ENSRNOT00000050702
|
Cdhr4
|
cadherin-related family member 4 |
| chr8_+_115150514 | 7.05 |
ENSRNOT00000016504
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr16_-_64806050 | 7.05 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr4_-_82263117 | 7.04 |
ENSRNOT00000008542
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr2_-_154418920 | 6.99 |
ENSRNOT00000076326
|
Plch1
|
phospholipase C, eta 1 |
| chr3_-_138462063 | 6.86 |
ENSRNOT00000065553
|
Ovol2
|
ovo-like zinc finger 2 |
| chr10_+_13736313 | 6.84 |
ENSRNOT00000079700
|
Abca3
|
ATP binding cassette subfamily A member 3 |
| chr10_+_56381813 | 6.72 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr14_-_103321270 | 6.68 |
ENSRNOT00000006157
|
Meis1
|
Meis homeobox 1 |
| chr17_+_47721977 | 6.57 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr3_-_2689084 | 6.50 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr19_-_56677084 | 6.48 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr4_+_44597123 | 6.45 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr8_-_115140080 | 6.45 |
ENSRNOT00000015851
|
Acy1
|
aminoacylase 1 |
| chr1_+_201055644 | 6.36 |
ENSRNOT00000054937
ENSRNOT00000047161 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr5_-_126911520 | 6.22 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr19_+_23389375 | 6.21 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr13_+_44424689 | 6.14 |
ENSRNOT00000005206
|
Acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chrX_+_110818716 | 6.10 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr7_-_11001933 | 6.09 |
ENSRNOT00000084006
|
Tle2
|
transducin-like enhancer of split 2 |
| chr6_+_107245820 | 5.89 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_-_221281180 | 5.80 |
ENSRNOT00000028379
|
Cdc42ep2
|
CDC42 effector protein 2 |
| chr17_-_14627937 | 5.66 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr17_-_88037034 | 5.65 |
ENSRNOT00000080193
ENSRNOT00000034907 |
Prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr17_+_5513930 | 5.56 |
ENSRNOT00000091370
|
Agtpbp1
|
ATP/GTP binding protein 1 |
| chr4_-_52350624 | 5.56 |
ENSRNOT00000060476
|
Tmem229a
|
transmembrane protein 229A |
| chr4_+_144382945 | 5.42 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr10_+_86950557 | 5.31 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr19_+_10563423 | 5.28 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr3_-_3574787 | 5.24 |
ENSRNOT00000087651
|
Nacc2
|
NACC family member 2 |
| chr3_+_11629556 | 5.21 |
ENSRNOT00000074401
|
St6galnac6
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chrX_+_156909913 | 5.19 |
ENSRNOT00000084390
|
L1cam
|
L1 cell adhesion molecule |
| chr2_-_102871257 | 5.16 |
ENSRNOT00000013116
|
Cyp7b1
|
cytochrome P450, family 7, subfamily b, polypeptide 1 |
| chr1_+_83653234 | 5.15 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr2_-_210726179 | 5.14 |
ENSRNOT00000025689
|
Gstm7
|
glutathione S-transferase, mu 7 |
| chr2_+_226900619 | 5.13 |
ENSRNOT00000019638
|
Pde5a
|
phosphodiesterase 5A |
| chr4_+_147692028 | 5.07 |
ENSRNOT00000014207
|
Cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr19_-_15421404 | 5.04 |
ENSRNOT00000083684
|
Slc6a2
|
solute carrier family 6 member 2 |
| chrX_+_68771100 | 4.99 |
ENSRNOT00000043872
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr4_+_31534225 | 4.88 |
ENSRNOT00000089292
ENSRNOT00000016423 |
NEWGENE_621351
|
collagen, type I, alpha 2 |
| chr4_+_7355574 | 4.78 |
ENSRNOT00000013800
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr3_+_122836446 | 4.65 |
ENSRNOT00000028816
|
Ebf4
|
early B-cell factor 4 |
| chr20_-_28814636 | 4.61 |
ENSRNOT00000086030
|
Sept10
|
septin 10 |
| chr3_+_145032200 | 4.54 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr5_+_76812931 | 4.51 |
ENSRNOT00000059458
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr8_+_85355766 | 4.50 |
ENSRNOT00000010583
|
Gcm1
|
glial cells missing homolog 1 |
| chr7_+_119647375 | 4.40 |
ENSRNOT00000046563
|
Kctd17
|
potassium channel tetramerization domain containing 17 |
| chr4_+_66090681 | 4.38 |
ENSRNOT00000078066
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr17_+_80882666 | 4.36 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr13_-_100672731 | 4.25 |
ENSRNOT00000004319
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr2_-_154508877 | 4.24 |
ENSRNOT00000086472
|
RGD1565059
|
similar to hypothetical protein E130311K13 |
| chr19_-_10596851 | 4.20 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr12_+_41543696 | 4.08 |
ENSRNOT00000079639
|
Tpcn1
|
two pore segment channel 1 |
| chr2_+_93792601 | 4.07 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr10_-_85084850 | 4.04 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr7_+_142575672 | 3.98 |
ENSRNOT00000080923
ENSRNOT00000008160 |
Scn8a
|
sodium voltage-gated channel alpha subunit 8 |
| chr4_+_7377563 | 3.93 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr16_-_6404578 | 3.89 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_-_187622373 | 3.89 |
ENSRNOT00000026396
|
Rhbg
|
Rh family B glycoprotein |
| chr2_+_184600721 | 3.85 |
ENSRNOT00000075823
|
Gatb
|
glutamyl-tRNA amidotransferase subunit B |
| chr1_+_221099998 | 3.84 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr10_+_65586504 | 3.82 |
ENSRNOT00000015535
ENSRNOT00000046388 |
Aldoc
|
aldolase, fructose-bisphosphate C |
| chr20_-_20680675 | 3.80 |
ENSRNOT00000087085
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr20_+_28815039 | 3.76 |
ENSRNOT00000075800
|
Sowahc
|
sosondowah ankyrin repeat domain family member C |
| chrX_-_82743753 | 3.72 |
ENSRNOT00000003512
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr8_-_60830448 | 3.65 |
ENSRNOT00000022419
|
Tspan3
|
tetraspanin 3 |
| chr13_+_79378733 | 3.59 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr1_-_49844547 | 3.58 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr10_+_57322331 | 3.57 |
ENSRNOT00000041493
|
Kif1c
|
kinesin family member 1C |
| chr10_-_70646495 | 3.56 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr15_-_27815261 | 3.54 |
ENSRNOT00000032992
|
Klhl33
|
kelch-like family member 33 |
| chr18_+_79406381 | 3.49 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr1_-_212021567 | 3.47 |
ENSRNOT00000029027
|
Cfap46
|
cilia and flagella associated protein 46 |
| chr14_-_8510138 | 3.33 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_+_61332351 | 3.26 |
ENSRNOT00000014492
|
Nrp1
|
neuropilin 1 |
| chr3_+_109862117 | 3.23 |
ENSRNOT00000083351
ENSRNOT00000070912 |
Thbs1
|
thrombospondin 1 |
| chr5_-_172424081 | 3.15 |
ENSRNOT00000019096
|
Plch2
|
phospholipase C, eta 2 |
| chr19_+_43163562 | 3.10 |
ENSRNOT00000048900
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr14_+_83724933 | 3.08 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr6_-_62798384 | 3.01 |
ENSRNOT00000093200
|
Ndufa10l1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10-like 1 |
| chr12_-_37538403 | 3.00 |
ENSRNOT00000001402
|
Snrnp35
|
small nuclear ribonucleoprotein U11/U12 subunit 35 |
| chr1_-_198104109 | 2.99 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr16_+_11599753 | 2.98 |
ENSRNOT00000079833
|
Grid1
|
glutamate ionotropic receptor delta type subunit 1 |
| chr19_+_43163129 | 2.93 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr1_+_135083974 | 2.89 |
ENSRNOT00000016481
ENSRNOT00000076098 |
Fam174b
|
family with sequence similarity 174, member B |
| chr12_+_40553741 | 2.87 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr13_-_72890841 | 2.82 |
ENSRNOT00000093726
|
RGD1304622
|
similar to 6820428L09 protein |
| chr1_-_184188911 | 2.80 |
ENSRNOT00000055124
ENSRNOT00000014948 |
Calca
|
calcitonin-related polypeptide alpha |
| chr7_-_144993652 | 2.77 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr13_+_42220251 | 2.67 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr4_-_117767772 | 2.66 |
ENSRNOT00000084170
|
LOC103690120
|
probable N-acetyltransferase CML1 |
| chrX_-_10218583 | 2.65 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr1_-_101351879 | 2.63 |
ENSRNOT00000028142
|
Ppfia3
|
PTPRF interacting protein alpha 3 |
| chr4_+_78233332 | 2.60 |
ENSRNOT00000011032
|
Repin1
|
replication initiator 1 |
| chr1_+_184299250 | 2.59 |
ENSRNOT00000030540
|
Insc
|
inscuteable homolog (Drosophila) |
| chr14_-_89208813 | 2.57 |
ENSRNOT00000091671
ENSRNOT00000006852 |
Hus1
|
HUS1 checkpoint clamp component |
| chr1_+_91152635 | 2.57 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr4_-_82263302 | 2.54 |
ENSRNOT00000082933
|
LOC100909604
|
homeobox protein Hox-A6-like |
| chr11_+_31002451 | 2.53 |
ENSRNOT00000002838
|
Eva1c
|
eva-1 homolog C |
| chr7_-_119441487 | 2.52 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr17_-_54714914 | 2.52 |
ENSRNOT00000024336
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr20_+_1749716 | 2.52 |
ENSRNOT00000048856
|
Olr1735
|
olfactory receptor 1735 |
| chr10_-_107376645 | 2.48 |
ENSRNOT00000046213
|
Cep295nl
|
CEP295 N-terminal like |
| chr1_+_257076221 | 2.43 |
ENSRNOT00000044895
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr1_-_219144610 | 2.42 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr10_-_74376270 | 2.41 |
ENSRNOT00000079311
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr4_+_145342193 | 2.39 |
ENSRNOT00000012015
|
Ttll3
|
tubulin tyrosine ligase like 3 |
| chr8_-_21995806 | 2.38 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr17_-_61332391 | 2.33 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr5_-_131860637 | 2.31 |
ENSRNOT00000064569
ENSRNOT00000080242 |
Slc5a9
|
solute carrier family 5 member 9 |
| chr10_-_56289882 | 2.25 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr12_-_51250230 | 2.24 |
ENSRNOT00000029508
|
Mn1
|
meningioma 1 |
| chr19_+_56026106 | 2.18 |
ENSRNOT00000066552
|
Cdk10
|
cyclin-dependent kinase 10 |
| chr14_+_46001849 | 2.13 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr1_-_71399751 | 2.12 |
ENSRNOT00000020787
|
Zfp444
|
zinc finger protein 444 |
| chr11_+_30363280 | 2.12 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr6_-_77421286 | 2.06 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr1_+_170383682 | 2.02 |
ENSRNOT00000024224
|
Smpd1
|
sphingomyelin phosphodiesterase 1 |
| chrX_+_122724129 | 2.00 |
ENSRNOT00000017746
|
LOC100360218
|
interleukin 13 receptor, alpha 1-like |
| chr6_+_127941526 | 1.97 |
ENSRNOT00000033897
|
LOC500712
|
Ab1-233 |
| chr8_+_85413537 | 1.96 |
ENSRNOT00000078159
|
Ick
|
intestinal cell kinase |
| chr1_+_266781617 | 1.94 |
ENSRNOT00000027417
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr9_+_46996156 | 1.91 |
ENSRNOT00000019673
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr11_+_69484293 | 1.90 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr4_-_117575154 | 1.88 |
ENSRNOT00000075813
|
LOC102556148
|
probable N-acetyltransferase CML2-like |
| chr7_+_116357783 | 1.86 |
ENSRNOT00000076279
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr2_+_198726118 | 1.83 |
ENSRNOT00000056237
|
Lix1l
|
limb and CNS expressed 1 like |
| chrX_+_29525256 | 1.75 |
ENSRNOT00000050018
|
Rab9a
|
RAB9A, member RAS oncogene family |
| chr7_-_123088819 | 1.74 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr20_+_27954433 | 1.73 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr1_+_215214853 | 1.69 |
ENSRNOT00000071235
ENSRNOT00000080442 |
LOC103690160
|
SH3 and multiple ankyrin repeat domains protein 2-like |
| chr5_+_137546860 | 1.63 |
ENSRNOT00000074431
|
Olr858
|
olfactory receptor 858 |
| chr5_-_173098816 | 1.62 |
ENSRNOT00000023696
|
Mib2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr5_+_140888973 | 1.61 |
ENSRNOT00000020524
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr8_-_22542997 | 1.58 |
ENSRNOT00000061009
|
Tmed1
|
transmembrane p24 trafficking protein 1 |
| chr9_+_82596355 | 1.57 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr1_-_175796040 | 1.56 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr1_-_100530183 | 1.55 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr1_-_89543967 | 1.48 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr17_+_85364483 | 1.44 |
ENSRNOT00000064257
|
Bmi1
|
BMI1 proto-oncogene, polycomb ring finger |
| chr13_-_90832469 | 1.42 |
ENSRNOT00000086508
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr9_-_82008620 | 1.41 |
ENSRNOT00000023365
|
Prkag3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chr5_-_171459488 | 1.37 |
ENSRNOT00000075423
ENSRNOT00000080686 |
Tprg1l
|
tumor protein p63 regulated 1-like |
| chr3_+_3788583 | 1.33 |
ENSRNOT00000084567
|
Gpsm1
|
G-protein signaling modulator 1 |
| chr10_+_14727180 | 1.32 |
ENSRNOT00000025236
|
Tpsg1
|
tryptase gamma 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.7 | 44.2 | GO:1901258 | mesangial cell-matrix adhesion(GO:0035759) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 5.6 | 22.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 3.7 | 15.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 3.7 | 25.8 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 2.9 | 8.7 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 2.6 | 7.8 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 2.6 | 26.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 2.5 | 7.5 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 2.5 | 7.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 2.3 | 13.9 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 2.3 | 6.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 2.1 | 6.4 | GO:2000286 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) regulation of inward rectifier potassium channel activity(GO:1901979) positive regulation of gap junction assembly(GO:1903598) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 2.1 | 8.3 | GO:0015746 | citrate transport(GO:0015746) |
| 2.1 | 6.2 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 1.9 | 5.6 | GO:0046038 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 1.9 | 18.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 1.8 | 12.8 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 1.8 | 5.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 1.8 | 14.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.7 | 24.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 1.7 | 5.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.7 | 5.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.6 | 6.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.5 | 15.4 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.4 | 8.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 1.3 | 5.2 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 1.3 | 3.9 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.3 | 3.9 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 1.3 | 5.1 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 1.3 | 3.8 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.2 | 3.6 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 1.2 | 3.5 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 1.2 | 9.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 1.1 | 4.4 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 1.1 | 3.3 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.1 | 3.2 | GO:0010752 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of cGMP-mediated signaling(GO:0010752) |
| 1.1 | 7.4 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 1.0 | 5.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.0 | 6.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 1.0 | 10.0 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.9 | 2.8 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.9 | 5.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.9 | 4.5 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.8 | 2.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.8 | 2.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.7 | 5.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.7 | 5.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.7 | 19.6 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.6 | 2.6 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.6 | 2.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.6 | 1.9 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.6 | 7.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.6 | 11.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.6 | 15.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 | 1.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.5 | 4.5 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.5 | 2.0 | GO:0035772 | interleukin-13-mediated signaling pathway(GO:0035772) |
| 0.5 | 3.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.5 | 1.5 | GO:0034769 | basement membrane disassembly(GO:0034769) pilomotor reflex(GO:0097195) |
| 0.5 | 2.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.5 | 5.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.5 | 1.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 11.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.4 | 3.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 | 6.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.4 | 1.3 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) ketone body catabolic process(GO:0046952) metanephric proximal tubule development(GO:0072237) |
| 0.4 | 3.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.4 | 2.4 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.4 | 2.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 5.0 | GO:0051934 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) |
| 0.3 | 5.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 12.1 | GO:0071526 | negative regulation of cell-matrix adhesion(GO:0001953) semaphorin-plexin signaling pathway(GO:0071526) |
| 0.3 | 1.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.3 | 2.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 3.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.3 | 3.7 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.3 | 2.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.3 | 1.0 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.3 | 6.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 4.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.3 | 2.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 1.8 | GO:0052428 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.3 | 1.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 1.4 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.3 | 2.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 13.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.3 | 3.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 4.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 8.8 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.3 | 16.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.3 | 8.5 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.2 | 33.9 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.2 | 4.7 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 1.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.2 | 1.5 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.2 | 1.6 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) |
| 0.2 | 0.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 8.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.2 | 8.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.7 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.2 | 4.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.2 | 4.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 2.6 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 0.7 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 4.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.9 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.1 | 1.3 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 12.3 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.1 | 3.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 6.2 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 2.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 2.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 5.7 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 3.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 9.4 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 5.4 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 3.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 3.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 10.8 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 2.6 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.1 | 6.1 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 7.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.0 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) |
| 0.1 | 0.7 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 1.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 1.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 5.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 3.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 4.6 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 1.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 1.0 | GO:0031023 | microtubule organizing center organization(GO:0031023) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 1.4 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.5 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 1.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 2.3 | 6.8 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 2.2 | 8.8 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 1.4 | 9.7 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 1.3 | 3.9 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 1.2 | 13.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.1 | 9.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 1.0 | 35.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.9 | 4.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.7 | 6.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.6 | 3.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 5.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 2.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 3.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.4 | 3.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 3.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 12.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 3.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.4 | 8.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 3.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 33.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 6.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 5.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 6.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 2.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 4.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 4.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 17.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 2.0 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 7.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 46.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 5.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 6.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 2.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 2.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 6.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 4.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 5.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 40.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 18.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 5.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 5.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 18.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 3.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 40.4 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 45.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 5.5 | 44.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 2.8 | 13.9 | GO:0031802 | type 1 metabotropic glutamate receptor binding(GO:0031798) type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 2.6 | 7.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 2.5 | 7.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 2.4 | 7.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 2.3 | 7.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 2.3 | 6.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 2.2 | 26.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 2.1 | 6.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 2.1 | 8.3 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 1.9 | 7.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.8 | 25.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.7 | 5.2 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 1.7 | 15.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.4 | 4.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 1.4 | 25.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.3 | 3.9 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 1.3 | 5.0 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 1.2 | 8.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.1 | 11.9 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 1.1 | 12.8 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.0 | 5.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.0 | 12.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.0 | 3.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.0 | 5.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.9 | 8.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.9 | 12.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.9 | 4.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.8 | 7.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.8 | 4.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.8 | 2.4 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.8 | 11.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.8 | 6.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 3.9 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.6 | 3.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 1.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.6 | 8.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 3.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.3 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.4 | 2.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 3.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 0.4 | 5.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 5.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 1.4 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.3 | 11.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.3 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 6.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 3.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 4.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 6.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 2.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 2.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 7.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 7.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 5.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 10.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 5.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 3.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 22.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 3.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 0.7 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.2 | 0.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.2 | 3.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 5.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 4.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.1 | 5.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 3.0 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 10.3 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 5.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 8.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 2.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 12.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 12.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 4.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 2.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 4.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.7 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 4.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 7.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 4.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 6.4 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 1.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 22.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 6.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 9.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 3.9 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 23.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 11.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 3.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 24.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 8.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 5.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 2.8 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 26.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 6.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 2.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 1.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 4.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 9.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 4.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 23.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 7.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 8.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 9.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 6.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 48.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.8 | 24.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.8 | 12.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 15.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.7 | 36.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.7 | 6.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 7.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.6 | 8.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 7.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 6.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 6.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 6.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 3.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 13.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 9.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 7.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 14.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.2 | 1.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 5.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 17.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 6.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 6.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 4.3 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 3.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.6 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 5.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 3.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |