Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
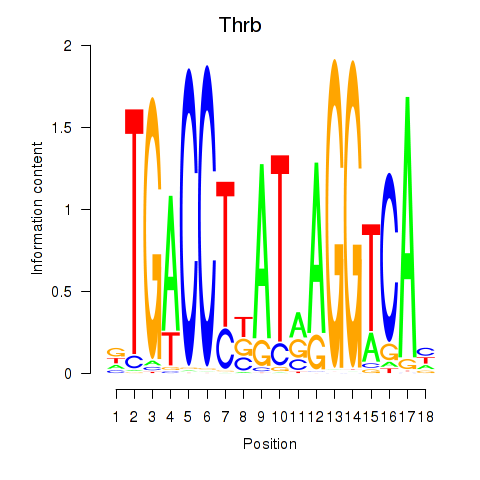
Results for Thrb
Z-value: 0.69
Transcription factors associated with Thrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thrb
|
ENSRNOG00000006649 | thyroid hormone receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thrb | rn6_v1_chr15_-_8914501_8914501 | -0.48 | 2.0e-19 | Click! |
Activity profile of Thrb motif
Sorted Z-values of Thrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_30276372 | 39.72 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr1_-_170431073 | 33.77 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr15_+_57290849 | 28.08 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr16_+_74865516 | 19.44 |
ENSRNOT00000058072
|
Atp7b
|
ATPase copper transporting beta |
| chr6_+_139486775 | 17.96 |
ENSRNOT00000077771
|
AABR07065699.3
|
|
| chr5_+_133865331 | 16.55 |
ENSRNOT00000035409
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr3_-_6626284 | 15.94 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr1_+_201660042 | 14.11 |
ENSRNOT00000093289
ENSRNOT00000054910 |
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr4_+_102147211 | 13.62 |
ENSRNOT00000083239
|
AABR07060980.1
|
|
| chr7_+_118712035 | 13.33 |
ENSRNOT00000074245
|
LOC100911562
|
apolipoprotein L3-like |
| chr1_+_88875375 | 13.23 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr20_-_5123073 | 12.55 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr17_+_81352700 | 12.43 |
ENSRNOT00000024736
|
Mrc1
|
mannose receptor, C type 1 |
| chr10_-_56962161 | 11.71 |
ENSRNOT00000026038
|
Alox15
|
arachidonate 15-lipoxygenase |
| chr1_+_219144205 | 11.62 |
ENSRNOT00000083942
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr5_+_34040258 | 11.15 |
ENSRNOT00000009758
|
Ggh
|
gamma-glutamyl hydrolase |
| chr3_-_4055806 | 11.15 |
ENSRNOT00000026408
|
Agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr9_+_14513037 | 9.94 |
ENSRNOT00000073130
|
Tspo2
|
translocator protein 2 |
| chr4_-_157252565 | 9.83 |
ENSRNOT00000079947
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_-_157252104 | 9.56 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr3_+_148438939 | 7.64 |
ENSRNOT00000064196
|
Ttll9
|
tubulin tyrosine ligase like 9 |
| chr2_-_88135410 | 7.62 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr8_+_122789453 | 7.35 |
ENSRNOT00000014585
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr3_-_82368357 | 7.02 |
ENSRNOT00000000052
|
Cd82
|
Cd82 molecule |
| chr2_+_127686925 | 6.18 |
ENSRNOT00000086653
ENSRNOT00000016946 |
Plk4
|
polo-like kinase 4 |
| chr11_+_85633243 | 6.02 |
ENSRNOT00000045807
|
LOC682352
|
Ig lambda chain V-VI region AR-like |
| chr20_-_2103864 | 5.89 |
ENSRNOT00000001014
|
Rnf39
|
ring finger protein 39 |
| chr10_+_31508512 | 5.81 |
ENSRNOT00000031580
|
Fam71b
|
family with sequence similarity 71, member B |
| chr13_+_50977431 | 5.72 |
ENSRNOT00000035658
|
Chit1
|
chitinase 1 |
| chr8_+_128027958 | 5.47 |
ENSRNOT00000045049
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr17_-_90522091 | 5.40 |
ENSRNOT00000077767
|
Lyst
|
lysosomal trafficking regulator |
| chr13_+_93751759 | 5.35 |
ENSRNOT00000065706
|
Wdr64
|
WD repeat domain 64 |
| chr5_-_19559244 | 5.19 |
ENSRNOT00000014289
ENSRNOT00000089666 |
Nsmaf
|
neutral sphingomyelinase activation associated factor |
| chr3_+_81294275 | 5.11 |
ENSRNOT00000009068
|
RGD1563263
|
similar to RIKEN cDNA 1700029I15 |
| chr10_+_108387789 | 4.99 |
ENSRNOT00000084845
|
AABR07030901.1
|
|
| chr2_-_210703265 | 4.87 |
ENSRNOT00000080228
|
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr9_+_17728816 | 4.58 |
ENSRNOT00000065754
|
Capn11
|
calpain 11 |
| chr4_+_174181644 | 4.57 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr10_+_92245442 | 4.26 |
ENSRNOT00000006808
|
Sppl2c
|
signal peptide peptidase like 2C |
| chr16_-_10706073 | 4.23 |
ENSRNOT00000089114
|
Fam25a
|
family with sequence similarity 25, member A |
| chr8_+_112594691 | 3.86 |
ENSRNOT00000038383
ENSRNOT00000081281 |
Acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr5_+_156668712 | 3.82 |
ENSRNOT00000067228
|
Ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase non-catalytic subunit |
| chr14_-_81053905 | 3.73 |
ENSRNOT00000045068
ENSRNOT00000040215 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr8_+_2635851 | 3.65 |
ENSRNOT00000061973
|
Casp4
|
caspase 4 |
| chr14_+_43694183 | 3.58 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr9_+_94310921 | 3.04 |
ENSRNOT00000026646
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr17_+_45175121 | 2.89 |
ENSRNOT00000080417
|
Nkapl
|
NFKB activating protein-like |
| chr16_-_21473808 | 2.86 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chr5_-_151117042 | 2.74 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr10_+_49368314 | 2.69 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chrX_+_124268129 | 2.53 |
ENSRNOT00000035489
|
Rhox13
|
Reproductive homeobox 13 |
| chrX_-_61151601 | 2.47 |
ENSRNOT00000004132
|
Mageb4
|
melanoma antigen family B, 4 |
| chr7_+_141053876 | 2.47 |
ENSRNOT00000084674
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_+_52147641 | 2.45 |
ENSRNOT00000009458
|
Hyal4
|
hyaluronoglucosaminidase 4 |
| chr12_+_50315933 | 2.31 |
ENSRNOT00000000826
|
Srrd
|
SRR1 domain containing |
| chr5_-_159371313 | 2.24 |
ENSRNOT00000009921
|
Padi1
|
peptidyl arginine deiminase 1 |
| chr17_-_22678253 | 2.07 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr3_-_7051953 | 2.03 |
ENSRNOT00000013473
|
RGD1306233
|
similar to hypothetical protein MGC29761 |
| chr4_+_72221550 | 1.80 |
ENSRNOT00000078533
|
Olr806
|
olfactory receptor 806 |
| chr4_-_56786754 | 1.70 |
ENSRNOT00000050795
|
Kcp
|
kielin/chordin-like protein |
| chr9_+_94310469 | 1.61 |
ENSRNOT00000065743
|
Eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chrX_-_102039530 | 1.54 |
ENSRNOT00000085348
|
AABR07040480.1
|
|
| chr3_-_119405453 | 1.49 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr10_-_110431792 | 1.13 |
ENSRNOT00000054922
|
LOC619574
|
hypothetical protein LOC619574 |
| chr13_+_109909053 | 0.89 |
ENSRNOT00000090541
|
AC125873.1
|
|
| chr15_-_27499998 | 0.79 |
ENSRNOT00000050189
|
LOC103693822
|
olfactory receptor 11H4-like |
| chr8_-_115358046 | 0.68 |
ENSRNOT00000017607
|
Grm2
|
glutamate metabotropic receptor 2 |
| chr13_+_91768256 | 0.64 |
ENSRNOT00000071206
|
LOC681470
|
similar to olfactory receptor 1403 |
| chr15_-_27485639 | 0.61 |
ENSRNOT00000041030
|
Olr1631
|
olfactory receptor 1631 |
| chr10_-_76263866 | 0.43 |
ENSRNOT00000003219
|
Scpep1
|
serine carboxypeptidase 1 |
| chr1_-_73733788 | 0.42 |
ENSRNOT00000025338
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr10_-_12386222 | 0.42 |
ENSRNOT00000060994
|
Olr1364
|
olfactory receptor 1364 |
| chr8_+_115131367 | 0.41 |
ENSRNOT00000014849
|
Rpl29
|
ribosomal protein L29 |
| chr7_-_144960527 | 0.36 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr3_-_20853708 | 0.35 |
ENSRNOT00000010360
|
Olr411
|
olfactory receptor 411 |
| chr15_-_27549748 | 0.35 |
ENSRNOT00000040096
|
Olr1635
|
olfactory receptor 1635 |
| chr10_-_44082313 | 0.34 |
ENSRNOT00000051126
|
Olr1421
|
olfactory receptor 1421 |
| chr8_-_111107599 | 0.30 |
ENSRNOT00000031313
|
Cep63
|
centrosomal protein 63 |
| chr3_-_160922341 | 0.21 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr10_+_44933574 | 0.09 |
ENSRNOT00000072854
|
LOC103690280
|
olfactory receptor 2AK2-like |
| chr10_+_44958544 | 0.01 |
ENSRNOT00000072282
|
Olr1439
|
olfactory receptor 1439 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 52.3 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 9.4 | 28.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 8.4 | 33.8 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 6.5 | 19.4 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 5.5 | 16.6 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 4.8 | 19.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 4.0 | 15.9 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 2.7 | 5.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 2.3 | 11.7 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 2.1 | 6.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 1.7 | 11.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) positive regulation of interleukin-12 secretion(GO:2001184) |
| 1.5 | 13.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 1.1 | 5.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.8 | 2.5 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.8 | 11.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.7 | 14.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.6 | 3.7 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.5 | 5.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.5 | 3.7 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.4 | 2.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 11.2 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.4 | 3.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 12.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 7.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 4.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 2.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.5 | GO:0006026 | aminoglycan catabolic process(GO:0006026) |
| 0.1 | 3.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.4 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 5.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 1.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 4.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 11.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 5.5 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 2.9 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 52.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 2.4 | 19.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.3 | 15.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.2 | 6.2 | GO:0098536 | deuterosome(GO:0098536) |
| 1.1 | 11.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.8 | 5.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.7 | 14.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.7 | 3.7 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.7 | 19.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 4.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 33.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 4.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 3.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 16.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 2.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.2 | 11.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.7 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 11.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 8.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 12.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 4.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 34.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 15.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 5.4 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 39.7 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 3.9 | 19.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 3.8 | 33.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.9 | 11.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 2.2 | 19.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 2.0 | 14.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 1.7 | 5.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.3 | 3.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.1 | 15.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.0 | 28.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.0 | 11.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 1.0 | 7.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 1.0 | 5.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.9 | 11.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.8 | 11.6 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.8 | 5.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.6 | 12.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 2.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 2.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 16.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 3.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 3.8 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 2.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 1.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 2.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 0.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 4.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 7.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 4.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 12.6 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 3.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 4.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 13.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 1.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 7.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 13.3 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 4.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 19.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.2 | 11.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 21.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 7.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 15.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 32.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.9 | 11.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.3 | 11.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.3 | 3.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 3.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 6.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |